Disrupted structure and aberrant function of CHIP mediates the loss of motor and cognitive function in preclinical models of SCAR16
- PMID: 30222779
- PMCID: PMC6160236
- DOI: 10.1371/journal.pgen.1007664
Disrupted structure and aberrant function of CHIP mediates the loss of motor and cognitive function in preclinical models of SCAR16
Abstract
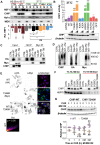
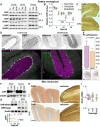
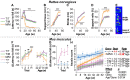
CHIP (carboxyl terminus of heat shock 70-interacting protein) has long been recognized as an active member of the cellular protein quality control system given the ability of CHIP to function as both a co-chaperone and ubiquitin ligase. We discovered a genetic disease, now known as spinocerebellar autosomal recessive 16 (SCAR16), resulting from a coding mutation that caused a loss of CHIP ubiquitin ligase function. The initial mutation describing SCAR16 was a missense mutation in the ubiquitin ligase domain of CHIP (p.T246M). Using multiple biophysical and cellular approaches, we demonstrated that T246M mutation results in structural disorganization and misfolding of the CHIP U-box domain, promoting oligomerization, and increased proteasome-dependent turnover. CHIP-T246M has no ligase activity, but maintains interactions with chaperones and chaperone-related functions. To establish preclinical models of SCAR16, we engineered T246M at the endogenous locus in both mice and rats. Animals homozygous for T246M had both cognitive and motor cerebellar dysfunction distinct from those observed in the CHIP null animal model, as well as deficits in learning and memory, reflective of the cognitive deficits reported in SCAR16 patients. We conclude that the T246M mutation is not equivalent to the total loss of CHIP, supporting the concept that disease-causing CHIP mutations have different biophysical and functional repercussions on CHIP function that may directly correlate to the spectrum of clinical phenotypes observed in SCAR16 patients. Our findings both further expand our basic understanding of CHIP biology and provide meaningful mechanistic insight underlying the molecular drivers of SCAR16 disease pathology, which may be used to inform the development of novel therapeutics for this devastating disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
The molecular basis of spinocerebellar ataxia type 48 caused by a de novo mutation in the ubiquitin ligase CHIP.J Biol Chem. 2022 May;298(5):101899. doi: 10.1016/j.jbc.2022.101899. Epub 2022 Apr 7. J Biol Chem. 2022. PMID: 35398354 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP.Hum Mol Genet. 2014 Feb 15;23(4):1013-24. doi: 10.1093/hmg/ddt497. Epub 2013 Oct 9. Hum Mol Genet. 2014. PMID: 24113144 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
A Crucial Role for the Protein Quality Control System in Motor Neuron Diseases.Front Aging Neurosci. 2020 Jul 21;12:191. doi: 10.3389/fnagi.2020.00191. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32792938 Free PMC article.
-
Imatinib induces ferroptosis in gastrointestinal stromal tumors by promoting STUB1-mediated GPX4 ubiquitination.Cell Death Dis. 2023 Dec 18;14(12):839. doi: 10.1038/s41419-023-06300-2. Cell Death Dis. 2023. PMID: 38110356 Free PMC article.
-
CHIP promotes Wnt signaling and regulates Arc stability by recruiting and polyubiquitinating LEF1 or Arc.Cell Death Discov. 2021 Jan 11;7(1):5. doi: 10.1038/s41420-020-00394-9. Cell Death Discov. 2021. PMID: 33431799 Free PMC article.
-
Die Hard: Necroptosis and its Impact on Age-Dependent Neuroinflammatory Diseases.Front Cell Death. 2024;3:1348153. doi: 10.3389/fceld.2024.1348153. Epub 2024 Mar 8. Front Cell Death. 2024. PMID: 40741039 Free PMC article.
-
CHIP mutations affect the heat shock response differently in human fibroblasts and iPSC-derived neurons.Dis Model Mech. 2020 Oct 12;13(10):dmm045096. doi: 10.1242/dmm.045096. Dis Model Mech. 2020. PMID: 33097556 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

