Genome-Wide Identification and Analysis of Apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER Family (NPF) Genes Reveals MdNPF6.5 Confers High Capacity for Nitrogen Uptake under Low-Nitrogen Conditions
- PMID: 30223432
- PMCID: PMC6164405
- DOI: 10.3390/ijms19092761
Genome-Wide Identification and Analysis of Apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER Family (NPF) Genes Reveals MdNPF6.5 Confers High Capacity for Nitrogen Uptake under Low-Nitrogen Conditions
Abstract
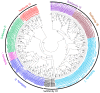
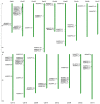
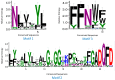
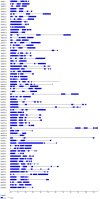
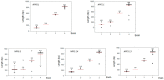
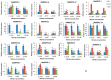
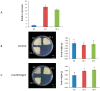
The NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) proteins play important roles in moving substrates such as nitrate, peptides, amino acids, dicarboxylates, malate, glucosinolates, indole acetic acid (IAA), abscisic acid (ABA), and jasmonic acid. Although a unified nomenclature of NPF members in plants has been reported, this gene family has not been studied as thoroughly in apple (Malus × domestica Borkh.) as it has in other species. Our objective was to provide general information about apple MdNPFs and analyze the transcriptional responses of some members to different levels of nitrate supplies. We identified 73 of these genes from the apple genome and used phylogenetic analysis to organize them into eight major groups. These apple NPFs are structurally conserved, based on alignment of amino acid sequences and analyses of phylogenetics and conserved domains. Examination of their genomic structures indicated that these genes are highly conserved among other species. We monitored 14 cloned MdNPFs that showed varied expression patterns under different nitrate concentrations and in different tissues. Among them, NPF6.5 was significantly induced by both low and high levels of nitrate. When compared with the wild type, 35S:MdNPF6.5 transgenic apple calli were more tolerant to low-N stress, which demonstrated that this gene confers greater capacity for nitrogen uptake under those conditions. We also analyzed the expression patterns of those 73 genes in various tissues. Our findings benefit future research on this family of genes.
Keywords: NPF gene family; apple; expression analysis; genome-wide; nitrate concentration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genome-wide analyses of genes encoding FK506-binding proteins reveal their involvement in abiotic stress responses in apple.BMC Genomics. 2018 Sep 25;19(1):707. doi: 10.1186/s12864-018-5097-8. BMC Genomics. 2018. PMID: 30253753 Free PMC article.
-
Genome-Wide Analysis and Cloning of the Apple Stress-Associated Protein Gene Family Reveals MdSAP15, Which Confers Tolerance to Drought and Osmotic Stresses in Transgenic Arabidopsis.Int J Mol Sci. 2018 Aug 21;19(9):2478. doi: 10.3390/ijms19092478. Int J Mol Sci. 2018. PMID: 30134640 Free PMC article.
-
A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants.Trends Plant Sci. 2014 Jan;19(1):5-9. doi: 10.1016/j.tplants.2013.08.008. Epub 2013 Sep 18. Trends Plant Sci. 2014. PMID: 24055139
-
Functional and Molecular Characterization of Plant Nitrate Transporters Belonging to NPF (NRT1/PTR) 6 Subfamily.Int J Mol Sci. 2024 Dec 20;25(24):13648. doi: 10.3390/ijms252413648. Int J Mol Sci. 2024. PMID: 39769409 Free PMC article. Review.
-
Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins.J Exp Bot. 2017 Jun 1;68(12):3107-3113. doi: 10.1093/jxb/erw499. J Exp Bot. 2017. PMID: 28186545 Review.
Cited by
-
Genome-wide identification and characterization of the NPF genes provide new insight into low nitrogen tolerance in Setaria.Front Plant Sci. 2022 Dec 14;13:1043832. doi: 10.3389/fpls.2022.1043832. eCollection 2022. Front Plant Sci. 2022. PMID: 36589108 Free PMC article.
-
MdMKK9-Mediated the Regulation of Anthocyanin Synthesis in Red-Fleshed Apple in Response to Different Nitrogen Signals.Int J Mol Sci. 2022 Jul 14;23(14):7755. doi: 10.3390/ijms23147755. Int J Mol Sci. 2022. PMID: 35887103 Free PMC article.
-
Genome-wide identification and expression analysis of the NRT genes in Ginkgo biloba under nitrate treatment reveal the potential roles during calluses browning.BMC Genomics. 2023 Oct 23;24(1):633. doi: 10.1186/s12864-023-09732-4. BMC Genomics. 2023. PMID: 37872493 Free PMC article.
-
Maize-soybean intercropping at optimal N fertilization increases the N uptake, N yield and N use efficiency of maize crop by regulating the N assimilatory enzymes.Front Plant Sci. 2023 Jan 4;13:1077948. doi: 10.3389/fpls.2022.1077948. eCollection 2022. Front Plant Sci. 2023. PMID: 36684768 Free PMC article.
-
Noncoding-RNA-Mediated Regulation in Response to Macronutrient Stress in Plants.Int J Mol Sci. 2021 Oct 18;22(20):11205. doi: 10.3390/ijms222011205. Int J Mol Sci. 2021. PMID: 34681864 Free PMC article. Review.
References
-
- Willmann A., Thomfohrde S., Haensch R., Nehls U. The poplar NRT2 gene family of high affinity nitrate importers: Impact of nitrogen nutrition and ectomycorrhiza formation. Environ. Exp. Bot. 2014;108:79–88. doi: 10.1016/j.envexpbot.2014.02.003. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

