TOP2B: The First Thirty Years
- PMID: 30223465
- PMCID: PMC6163646
- DOI: 10.3390/ijms19092765
TOP2B: The First Thirty Years
Abstract
Type II DNA topoisomerases (EC 5.99.1.3) are enzymes that catalyse topological changes in DNA in an ATP dependent manner. Strand passage reactions involve passing one double stranded DNA duplex (transported helix) through a transient enzyme-bridged break in another (gated helix). This activity is required for a range of cellular processes including transcription. Vertebrates have two isoforms: topoisomerase IIα and β. Topoisomerase IIβ was first reported in 1987. Here we review the research on DNA topoisomerase IIβ over the 30 years since its discovery.
Keywords: Cell cycle; TOP2; Topoisomerase II; cell biology; review; transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
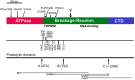

References
-
- Drake F.H., Zimmerman J.P., McCabe F.L., Bartus H.F., Per S.R., Sullivan D.M., Ross W.E., Mattern M.R., Johnson R.K., Crooke S.T. Purification of topoisomerase II from amsacrine-resistant P388 leukemia cells. Evidence for two forms of the enzyme. J. Biol. Chem. 1987;262:16739–16747. - PubMed
-
- Per S.R., Mattern M.R., Mirabelli C.K., Drake F.H., Johnson R.K., Crooke S.T. Characterization of a subline of P388 leukemia resistant to amsacrine: Evidence of altered topoisomerase II function. Mol. Pharmacol. 1987;32:17–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

