The Cytoscan HD Array in the Diagnosis of Neurodevelopmental Disorders
- PMID: 30223503
- PMCID: PMC6164295
- DOI: 10.3390/ht7030028
The Cytoscan HD Array in the Diagnosis of Neurodevelopmental Disorders
Abstract
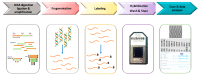
Submicroscopic chromosomal copy number variations (CNVs), such as deletions and duplications, account for about 15⁻20% of patients affected with developmental delay, intellectual disability, multiple congenital anomalies, and autism spectrum disorder. Most of CNVs are de novo or inherited rearrangements with clinical relevance, but there are also rare inherited imbalances with unknown significance that make difficult the clinical management and genetic counselling. Chromosomal microarrays analysis (CMA) are recognized as the first-line test for CNV detection and are now routinely used in the clinical diagnostic laboratory. The recent use of CMA platforms that combine classic copy number analysis with single-nucleotide polymorphism (SNP) genotyping has increased the diagnostic yields. Here we discuss the application of the Cytoscan high-density (HD) SNP-array for the detection of CNVs. We provide an overview of molecular analyses involved in identifying pathogenic CNVs and highlight important guidelines to establish pathogenicity of CNV.
Keywords: SNP-array; copy number variations; neurodevelopmental disorders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. - DOI - PMC - PubMed
-
- American Academy of Neurology Evidence Repot: Genetic and Metabolic Testing in Children with Global Developmental Delay. [(accessed on 10 July 2018)]; Available online: https://www.aan.com/Guidelines/home/GuidelineDetail/487.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

