Restriction enzyme digestion of host DNA enhances universal detection of parasitic pathogens in blood via targeted amplicon deep sequencing
- PMID: 30223888
- PMCID: PMC6142370
- DOI: 10.1186/s40168-018-0540-2
Restriction enzyme digestion of host DNA enhances universal detection of parasitic pathogens in blood via targeted amplicon deep sequencing
Abstract
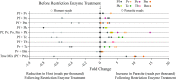
Background: Targeted amplicon deep sequencing (TADS) of the 16S rRNA gene is commonly used to explore and characterize bacterial microbiomes. Meanwhile, attempts to apply TADS to the detection and characterization of entire parasitic communities have been hampered since conserved regions of many conserved parasite genes, such as the 18S rRNA gene, are also conserved in their eukaryotic hosts. As a result, targeted amplification of 18S rRNA from clinical samples using universal primers frequently results in competitive priming and preferential amplification of host DNA. Here, we describe a novel method that employs a single pair of universal primers to capture all blood-borne parasites while reducing host 18S rRNA template and enhancing the amplification of parasite 18S rRNA for TADS. This was achieved using restriction enzymes to digest the 18S rRNA gene at cut sites present only in the host sequence prior to PCR amplification.
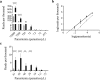
Results: This method was validated against 16 species of blood-borne helminths and protozoa. Enzyme digestion prior to PCR enrichment and Illumina amplicon deep sequencing led to a substantial reduction in human reads and a corresponding 5- to 10-fold increase in parasite reads relative to undigested samples. This method allowed for discrimination of all common parasitic agents found in human blood, even in cases of multi-parasite infection, and markedly reduced the limit of detection in digested versus undigested samples.
Conclusions: The results herein provide a novel methodology for the reduction of host DNA prior to TADS and establish the validity of a next-generation sequencing-based platform for universal parasite detection.
Keywords: Amplicon sequencing; Blood microbiota; Molecular parasitology; Parasite biodiversity.
Conflict of interest statement
Ethics approval and consent to participate
Ethics approval for the use of anonymized, de-identified, non-reidentifiable blood samples as non-engaged research was granted by Centers for Disease Control and Prevention Division of Parasitic Diseases and Malaria Human Subjects Review, approval number 2016-314. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry.
Competing interests
E.T., R.S.B., C.O., and B.R.F. have submitted a patent (E-113-2017/0; I-024-16) for the use of restriction enzymes to reduce host DNA in TADS analyses. The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Sensitive universal detection of blood parasites by selective pathogen-DNA enrichment and deep amplicon sequencing.Microbiome. 2021 Jan 2;9(1):1. doi: 10.1186/s40168-020-00939-1. Microbiome. 2021. PMID: 33388088 Free PMC article.
-
Simultaneous targeted amplicon deep sequencing and library preparation for a time and cost-effective universal parasite diagnostic sequencing approach.Parasitol Res. 2023 Dec;122(12):3243-3256. doi: 10.1007/s00436-023-07991-4. Epub 2023 Nov 9. Parasitol Res. 2023. PMID: 37940706 Free PMC article.
-
Untargeted metagenomics shows a reliable performance for synchronous detection of parasites.Parasitol Res. 2020 Aug;119(8):2623-2629. doi: 10.1007/s00436-020-06754-9. Epub 2020 Jun 26. Parasitol Res. 2020. PMID: 32591865 Free PMC article.
-
Molecular diagnosis in clinical parasitology: when and why?Exp Biol Med (Maywood). 2014 Nov;239(11):1443-60. doi: 10.1177/1535370214523880. Epub 2014 Mar 25. Exp Biol Med (Maywood). 2014. PMID: 24668556 Review.
-
[Molecular diagnostic of parasites using rRNA gene sequence].Wiad Parazytol. 2006;52(4):263-9. Wiad Parazytol. 2006. PMID: 17432616 Review. Polish.
Cited by
-
Considerations for mosquito microbiome research from the Mosquito Microbiome Consortium.Microbiome. 2021 Feb 1;9(1):36. doi: 10.1186/s40168-020-00987-7. Microbiome. 2021. PMID: 33522965 Free PMC article. Review.
-
Contrasting Strategies: Human Eukaryotic Versus Bacterial Microbiome Research.J Eukaryot Microbiol. 2020 Mar;67(2):279-295. doi: 10.1111/jeu.12766. Epub 2019 Oct 20. J Eukaryot Microbiol. 2020. PMID: 31583780 Free PMC article. Review.
-
Development of a Blocking Primer to Inhibit the PCR Amplification of the 18S rDNA Sequences of Litopenaeus vannamei and Its Efficacy in Crassostrea hongkongensis.Front Microbiol. 2019 Apr 23;10:830. doi: 10.3389/fmicb.2019.00830. eCollection 2019. Front Microbiol. 2019. PMID: 31065252 Free PMC article.
-
Nested qPCR assay to detect Babesia duncani infection in hamsters and humans.Parasitol Res. 2022 Dec;121(12):3603-3610. doi: 10.1007/s00436-022-07685-3. Epub 2022 Oct 4. Parasitol Res. 2022. PMID: 36192649
-
Where Have All the Diagnostic Morphological Parasitologists Gone?J Clin Microbiol. 2022 Nov 16;60(11):e0098622. doi: 10.1128/jcm.00986-22. Epub 2022 Oct 31. J Clin Microbiol. 2022. PMID: 36314793 Free PMC article.
References
-
- Tanaka Ryusei, Hino Akina, Tsai Isheng J., Palomares-Rius Juan Emilio, Yoshida Ayako, Ogura Yoshitoshi, Hayashi Tetsuya, Maruyama Haruhiko, Kikuchi Taisei. Assessment of Helminth Biodiversity in Wild Rats Using 18S rDNA Based Metagenomics. PLoS ONE. 2014;9(10):e110769. doi: 10.1371/journal.pone.0110769. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

