Surfing Motility: a Conserved yet Diverse Adaptation among Motile Bacteria
- PMID: 30224438
- PMCID: PMC6222206
- DOI: 10.1128/JB.00394-18
Surfing Motility: a Conserved yet Diverse Adaptation among Motile Bacteria
Abstract
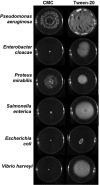
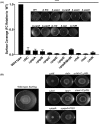
Bacterial rapid surfing motility is a novel surface adaptation of Pseudomonas aeruginosa in the presence of the glycoprotein mucin. Here, we show that other Gram-negative motile bacterial species, including Escherichia coli, Salmonella enterica, Vibrio harveyi, Enterobacter cloacae, and Proteus mirabilis, also exhibit the physical characteristics of surfing on the surface of agar plates containing 0.4% mucin, where surfing motility was generally more rapid and less dependent on medium viscosity than was swimming motility. As previously observed in Pseudomonas aeruginosa, all surfing species exhibited some level of broad-spectrum adaptive resistance, although the antibiotics to which they demonstrated surfing-mediated resistance differed. Surfing motility in P. aeruginosa was found to be dependent on the quorum-sensing systems of this organism; however, this aspect was not conserved in other tested bacterial species, including V. harveyi and S. enterica, as demonstrated by assaying specific quorum-sensing mutants. Thus, rapid surfing motility is a complex surface growth adaptation that is conserved in several motile bacteria, involves flagella, and leads to diverse broad-spectrum antibiotic resistance, but it is distinct in terms of dependence on quorum sensing.IMPORTANCE This study showed for the first time that surfing motility, a novel form of surface motility first discovered in Pseudomonas aeruginosa under artificial cystic fibrosis conditions, including the presence of high mucin content, is conserved in other motile bacterial species known to be mucosa-associated, including Escherichia coli, Salmonella enterica, and Proteus mirabilis Here, we demonstrated that key characteristics of surfing, including the ability to adapt to various viscous environments and multidrug adaptive resistance, are also conserved. Using mutagenesis assays, we also identified the importance of all three known quorum-sensing systems, Las, Rhl, and Pqs, in P. aeruginosa in regulating surfing motility, and we also observed a conserved dependence of surfing on flagella in certain species.
Keywords: antibiotic resistance; bacterial motility; mucin; quorum sensing; surface motility; surfing.
Copyright © 2018 American Society for Microbiology.
Figures




Comment in
-
Surf's up!Nat Rev Microbiol. 2018 Nov;16(11):658-659. doi: 10.1038/s41579-018-0093-1. Nat Rev Microbiol. 2018. PMID: 30262844 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

