Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells
- PMID: 30224477
- PMCID: PMC6176615
- DOI: 10.1073/pnas.1807704115
Control of CCND1 ubiquitylation by the catalytic SAGA subunit USP22 is essential for cell cycle progression through G1 in cancer cells
Abstract
Overexpression of the deubiquitylase ubiquitin-specific peptidase 22 (USP22) is a marker of aggressive cancer phenotypes like metastasis, therapy resistance, and poor survival. Functionally, this overexpression of USP22 actively contributes to tumorigenesis, as USP22 depletion blocks cancer cell cycle progression in vitro, and inhibits tumor progression in animal models of lung, breast, bladder, ovarian, and liver cancer, among others. Current models suggest that USP22 mediates these biological effects via its role in epigenetic regulation as a subunit of the Spt-Ada-Gcn5-acetyltransferase (SAGA) transcriptional cofactor complex. Challenging the dogma, we report here a nontranscriptional role for USP22 via a direct effect on the core cell cycle machinery: that is, the deubiquitylation of the G1 cyclin D1 (CCND1). Deubiquitylation by USP22 protects CCND1 from proteasome-mediated degradation and occurs separately from the canonical phosphorylation/ubiquitylation mechanism previously shown to regulate CCND1 stability. We demonstrate that control of CCND1 is a key mechanism by which USP22 mediates its known role in cell cycle progression. Finally, USP22 and CCND1 levels correlate in patient lung and colorectal cancer samples and our preclinical studies indicate that targeting USP22 in combination with CDK inhibitors may offer an approach for treating cancer patients whose tumors exhibit elevated CCND1.
Keywords: CCND1; SAGA; USP22; cell cycle; deubiquitylation.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: The authors declare no conflict of interest.
Figures
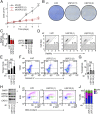

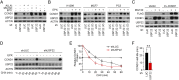



References
-
- Zhao Y, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. - PubMed
-
- Glinsky GV. Death-from-cancer signatures and stem cell contribution to metastatic cancer. Cell Cycle. 2005;4:1171–1175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

