Influenza hemagglutinin membrane anchor
- PMID: 30224494
- PMCID: PMC6176637
- DOI: 10.1073/pnas.1810927115
Influenza hemagglutinin membrane anchor
Abstract
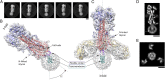
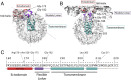
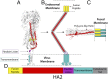
Viruses with membranes fuse them with cellular membranes, to transfer their genomes into cells at the beginning of infection. For Influenza virus, the membrane glycoprotein involved in fusion is the hemagglutinin (HA), the 3D structure of which is known from X-ray crystallographic studies. The soluble ectodomain fragments used in these studies lacked the "membrane anchor" portion of the molecule. Since this region has a role in membrane fusion, we have determined its structure by analyzing the intact, full-length molecule in a detergent micelle, using cryo-EM. We have also compared the structures of full-length HA-detergent micelles with full-length HA-Fab complex detergent micelles, to describe an infectivity-neutralizing monoclonal Fab that binds near the ectodomain membrane anchor junction. We determine a high-resolution HA structure which compares favorably in detail with the structure of the ectodomain seen by X-ray crystallography; we detect, clearly, all five carbohydrate side chains of HA; and we find that the ectodomain is joined to the membrane anchor by flexible, eight-residue-long, linkers. The linkers extend into the detergent micelle to join a central triple-helical structure that is a major component of the membrane anchor.
Keywords: cryo-EM; hemagglutinin; influenza; membrane fusion; membrane protein.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: A.L. is the scientific founder of Humabs BioMed SA. A.L. holds shares in Humabs BioMed. D.C. is an employee of Humabs Biomed. N.L.K. was employed by MedImmune, LLC when work was executed and may currently hold AstraZeneca stock or stock options.
Figures


References
-
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. - PubMed
-
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
-
- Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature. 1981;290:713–717. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

