sox9b is required in cardiomyocytes for cardiac morphogenesis and function
- PMID: 30224706
- PMCID: PMC6141582
- DOI: 10.1038/s41598-018-32125-7
sox9b is required in cardiomyocytes for cardiac morphogenesis and function
Abstract
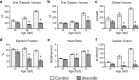
The high mobility group transcription factor SOX9 is expressed in stem cells, progenitor cells, and differentiated cell-types in developing and mature organs. Exposure to a variety of toxicants including dioxin, di(2-ethylhexyl) phthalate, 6:2 chlorinated polyfluorinated ether sulfonate, and chlorpyrifos results in the downregulation of tetrapod Sox9 and/or zebrafish sox9b. Disruption of Sox9/sox9b function through environmental exposures or genetic mutations produce a wide range of phenotypes and adversely affect organ development and health. We generated a dominant-negative sox9b (dnsox9b) to inhibit sox9b target gene expression and used the Gal4/UAS system to drive dnsox9b specifically in cardiomyocytes. Cardiomyocyte-specific inhibition of sox9b function resulted in a decrease in ventricular cardiomyocytes, an increase in atrial cardiomyocytes, hypoplastic endothelial cushions, and impaired epicardial development, ultimately culminating in heart failure. Cardiomyocyte-specific dnsox9b expression significantly reduced end diastolic volume, which corresponded with a decrease in stroke volume, ejection fraction, and cardiac output. Further analysis of isolated cardiac tissue by RT-qPCR revealed cardiomyocyte-specific inhibition of sox9b function significantly decreased the expression of the critical cardiac development genes nkx2.5, nkx2.7, and myl7, as well as c-fos, an immediate early gene necessary for cardiomyocyte progenitor differentiation. Together our studies indicate sox9b transcriptional regulation is necessary for cardiomyocyte development and function.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Jenkins KJ, et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

