Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4
- PMID: 30224758
- PMCID: PMC6181206
- DOI: 10.1038/s41591-018-0158-8
Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4
Abstract
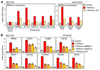
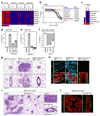
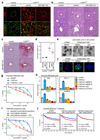
Cancer cells rely on dysregulated gene expression. This establishes specific transcriptional addictions that may be therapeutically exploited. Yet, the mechanisms that are ultimately responsible for these addictions are poorly understood. Here, we investigated the transcriptional dependencies of transformed cells to the transcription factors YAP and TAZ. YAP/TAZ physically engage the general coactivator bromodomain-containing protein 4 (BRD4), dictating the genome-wide association of BRD4 to chromatin. YAP/TAZ flag a large set of enhancers with super-enhancer-like functional properties. YAP/TAZ-bound enhancers mediate the recruitment of BRD4 and RNA polymerase II at YAP/TAZ-regulated promoters, boosting the expression of a host of growth-regulating genes. Treatment with small-molecule inhibitors of BRD4 blunts YAP/TAZ pro-tumorigenic activity in several cell or tissue contexts, causes the regression of pre-established, YAP/TAZ-addicted neoplastic lesions and reverts drug resistance. This work sheds light on essential mediators, mechanisms and genome-wide regulatory elements that are responsible for transcriptional addiction in cancer and lays the groundwork for a rational use of BET inhibitors according to YAP/TAZ biology.
Conflict of interest statement
Figures



Comment in
-
BETting on YAP-TAZ.Nat Rev Cancer. 2018 Nov;18(11):663. doi: 10.1038/s41568-018-0065-9. Nat Rev Cancer. 2018. PMID: 30301931 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

