Single particle trajectories reveal active endoplasmic reticulum luminal flow
- PMID: 30224760
- PMCID: PMC6435195
- DOI: 10.1038/s41556-018-0192-2
Single particle trajectories reveal active endoplasmic reticulum luminal flow
Abstract
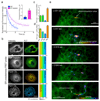
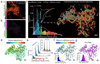
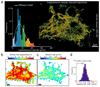
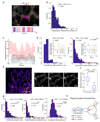
The endoplasmic reticulum (ER), a network of membranous sheets and pipes, supports functions encompassing biogenesis of secretory proteins and delivery of functional solutes throughout the cell1,2. Molecular mobility through the ER network enables these functionalities, but diffusion alone is not sufficient to explain luminal transport across supramicrometre distances. Understanding the ER structure-function relationship is critical in light of mutations in ER morphology-regulating proteins that give rise to neurodegenerative disorders3,4. Here, super-resolution microscopy and analysis of single particle trajectories of ER luminal proteins revealed that the topological organization of the ER correlates with distinct trafficking modes of its luminal content: with a dominant diffusive component in tubular junctions and a fast flow component in tubules. Particle trajectory orientations resolved over time revealed an alternating current of the ER contents, while fast ER super-resolution identified energy-dependent tubule contraction events at specific points as a plausible mechanism for generating active ER luminal flow. The discovery of active flow in the ER has implications for timely ER content distribution throughout the cell, particularly important for cells with extensive ER-containing projections such as neurons.
Figures

References
-
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. - PubMed
-
- Hubner CA, Kurth I. Membrane-shaping disorders: a common pathway in axon degeneration. Brain : a journal of neurology. 2014;137:3109–3121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/K015850/1/MRC_/Medical Research Council/United Kingdom
- MR/K02292X/1/MRC_/Medical Research Council/United Kingdom
- G0601840/MRC_/Medical Research Council/United Kingdom
- G1002610/MRC_/Medical Research Council/United Kingdom
- BB/H023917/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

