Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA
- PMID: 30224964
- PMCID: PMC6126647
- DOI: 10.1107/S205225251800996X
Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA
Abstract
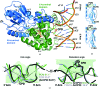
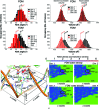
Cyclobutane pyrimidine dimer (CPD) photolyases harness the energy of blue light to repair UV-induced DNA CPDs. Upon binding, CPD photolyases cause the photodamage to flip out of the duplex DNA and into the catalytic site of the enzyme. This process, called base-flipping, induces a kink in the DNA, as well as an unpaired bubble, which are stabilized by a network of protein-nucleic acid interactions. Previously, several co-crystal structures have been reported in which the binding mode of CPD photolyases has been studied in detail. However, in all cases the internucleoside linkage of the photodamage site was a chemically synthesized formacetal analogue and not the natural phosphodiester. Here, the first crystal structure and conformational analysis via molecular-dynamics simulations of a class II CPD photolyase in complex with photodamaged DNA that contains a natural cyclobutane pyrimidine dimer with an intra-lesion phosphodiester linkage are presented. It is concluded that a highly conserved bubble-intruding region (BIR) mediates stabilization of the open form of CPD DNA when complexed with class II CPD photolyases.
Keywords: DNA distortion; DNA repair; class II CPD photolyases; crystal structure; cyclobutane pyrimidine dimer.
Figures

Similar articles
-
Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA.EMBO J. 2011 Sep 2;30(21):4437-49. doi: 10.1038/emboj.2011.313. EMBO J. 2011. PMID: 21892138 Free PMC article.
-
Influence of a cis,syn-cyclobutane pyrimidine dimer damage on DNA conformation studied by molecular dynamics simulations.Biopolymers. 2015 Apr;103(4):215-22. doi: 10.1002/bip.22586. Biopolymers. 2015. PMID: 25382106
-
Transport of rice cyclobutane pyrimidine dimer photolyase into mitochondria relies on a targeting sequence located in its C-terminal internal region.Plant J. 2014 Sep;79(6):951-63. doi: 10.1111/tpj.12598. Epub 2014 Jul 31. Plant J. 2014. PMID: 24947012
-
A Review of Spectroscopic and Biophysical-Chemical Studies of the Complex of Cyclobutane Pyrimidine Dimer Photolyase and Cryptochrome DASH with Substrate DNA.Photochem Photobiol. 2017 Jan;93(1):26-36. doi: 10.1111/php.12678. Epub 2017 Jan 20. Photochem Photobiol. 2017. PMID: 27891613 Review.
-
Photolyase: Dynamics and electron-transfer mechanisms of DNA repair.Arch Biochem Biophys. 2017 Oct 15;632:158-174. doi: 10.1016/j.abb.2017.08.007. Epub 2017 Aug 9. Arch Biochem Biophys. 2017. PMID: 28802828 Free PMC article. Review.
Cited by
-
Formation and Recognition of UV-Induced DNA Damage within Genome Complexity.Int J Mol Sci. 2020 Sep 12;21(18):6689. doi: 10.3390/ijms21186689. Int J Mol Sci. 2020. PMID: 32932704 Free PMC article. Review.
-
Theoretical insights into the DNA repair function of Arabidopsis thaliana cryptochrome-DASH.Biophys Physicobiol. 2020 Sep 4;17:113-124. doi: 10.2142/biophysico.BSJ-2020010. eCollection 2020. Biophys Physicobiol. 2020. PMID: 33194514 Free PMC article.
-
Light-driven DNA repair at atomic and picosecond resolution revealed via time-resolved serial femtosecond crystallography.Clin Transl Med. 2024 Jan;14(1):e1566. doi: 10.1002/ctm2.1566. Clin Transl Med. 2024. PMID: 38279893 Free PMC article. No abstract available.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

