The complete plastid genomes of Ophrys iricolor and O. sphegodes (Orchidaceae) and comparative analyses with other orchids
- PMID: 30226857
- PMCID: PMC6143245
- DOI: 10.1371/journal.pone.0204174
The complete plastid genomes of Ophrys iricolor and O. sphegodes (Orchidaceae) and comparative analyses with other orchids
Abstract
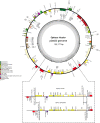
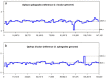
Sexually deceptive orchids of the genus Ophrys may rapidly evolve by adaptation to pollinators. However, understanding of the genetic basis of potential changes and patterns of relationships is hampered by a lack of genomic information. We report the complete plastid genome sequences of Ophrys iricolor and O. sphegodes, representing the two most species-rich lineages of the genus Ophrys. Both plastomes are circular DNA molecules (146754 bp for O. sphegodes and 150177 bp for O. iricolor) with the typical quadripartite structure of plastid genomes and within the average size of photosynthetic orchids. 213 Simple Sequence Repeats (SSRs) (31.5% polymorphic between O. iricolor and O. sphegodes) were identified, with homopolymers and dipolymers as the most common repeat types. SSRs were mainly located in intergenic regions but SSRs located in coding regions were also found, mainly in ycf1 and rpoC2 genes. The Ophrys plastome is predicted to encode 107 distinct genes, 17 of which are completely duplicated in the Inverted Repeat regions. 83 and 87 putative RNA editing sites were detected in 25 plastid genes of the two Ophrys species, all occurring in the first or second codon position. Comparing the rate of nonsynonymous (dN) and synonymous (dS) substitutions, 24 genes (including rbcL and ycf1) display signature consistent with positive selection. When compared with other members of the orchid family, the Ophrys plastome has a complete set of 11 functional ndh plastid genes, with the exception of O. sphegodes that has a truncated ndhF gene. Comparative analysis showed a large co-linearity with other related Orchidinae. However, in contrast to O. iricolor and other Orchidinae, O. sphegodes has a shift of the junction between the Inverted Repeat and Small Single Copy regions associated with the loss of the partial duplicated gene ycf1 and the truncation of the ndhF gene. Data on relative genomic coverage and validation by PCR indicate the presence, with a different ratio, of the two plastome types (i.e. with and without ndhF deletion) in both Ophrys species, with a predominance of the deleted type in O. sphegodes. A search for this deleted plastid region in O. sphegodes nuclear genome shows that the deleted region is inserted in a retrotransposon nuclear sequence. The present study provides useful genomic tools for studying conservation and patterns of relationships of this rapidly radiating orchid genus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications.PLoS One. 2014 Jun 9;9(6):e99016. doi: 10.1371/journal.pone.0099016. eCollection 2014. PLoS One. 2014. PMID: 24911363 Free PMC article.
-
Pollinator shifts between Ophrys sphegodes populations: might adaptation to different pollinators drive population divergence?J Evol Biol. 2013 Oct;26(10):2197-208. doi: 10.1111/jeb.12216. Epub 2013 Aug 28. J Evol Biol. 2013. PMID: 23981167
-
A Phylogenomic Analysis of the Floral Transcriptomes of Sexually Deceptive and Rewarding European Orchids, Ophrys and Gymnadenia.Front Plant Sci. 2019 Nov 29;10:1553. doi: 10.3389/fpls.2019.01553. eCollection 2019. Front Plant Sci. 2019. PMID: 31850034 Free PMC article.
-
Dynamic evolution of the plastome in the Elm family (Ulmaceae).Planta. 2022 Dec 17;257(1):14. doi: 10.1007/s00425-022-04045-4. Planta. 2022. PMID: 36526857 Review.
-
In-depth analysis of genomes and functional genomics of orchid using cutting-edge high-throughput sequencing.Front Plant Sci. 2022 Sep 23;13:1018029. doi: 10.3389/fpls.2022.1018029. eCollection 2022. Front Plant Sci. 2022. PMID: 36212315 Free PMC article. Review.
Cited by
-
The Complete Chloroplast Genome of Euphrasia regelii, Pseudogenization of ndh Genes and the Phylogenetic Relationships Within Orobanchaceae.Front Genet. 2019 May 14;10:444. doi: 10.3389/fgene.2019.00444. eCollection 2019. Front Genet. 2019. PMID: 31156705 Free PMC article.
-
Extremely low levels of chloroplast genome sequence variability in Astelia pumila (Asteliaceae, Asparagales).PeerJ. 2019 Jan 17;7:e6244. doi: 10.7717/peerj.6244. eCollection 2019. PeerJ. 2019. PMID: 30671303 Free PMC article.
-
Target Nuclear and Off-Target Plastid Hybrid Enrichment Data Inform a Range of Evolutionary Depths in the Orchid Genus Epidendrum.Front Plant Sci. 2020 Jan 29;10:1761. doi: 10.3389/fpls.2019.01761. eCollection 2019. Front Plant Sci. 2020. PMID: 32063915 Free PMC article.
-
Evolutionary differences in gene loss and pseudogenization among mycoheterotrophic orchids in the tribe Vanilleae (subfamily Vanilloideae).Front Plant Sci. 2023 Mar 22;14:1160446. doi: 10.3389/fpls.2023.1160446. eCollection 2023. Front Plant Sci. 2023. PMID: 37035052 Free PMC article.
-
Plastid phylogenomics reveals evolutionary relationships in the mycoheterotrophic orchid genus Dipodium and provides insights into plastid gene degeneration.Front Plant Sci. 2024 Jun 13;15:1388537. doi: 10.3389/fpls.2024.1388537. eCollection 2024. Front Plant Sci. 2024. PMID: 38938632 Free PMC article.
References
-
- Palmer JD (1991) Plastid chromosomes: structure and evolution In: Vasil LK, Bogorad L, editors. Cell Culture and Somatic Cell Genetics in Plants, the Molecular Biology of Plastid 7A. San Diego: Academic Press; pp. 5–53.
-
- Downie SR, Palmer JD (1992) Use of chloroplast DNA rearrangements in reconstructing plant phylogeny In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular Systematics of Plants. Springer; US; pp. 14–35.
-
- Judd WS, Campbell CS, Kellogg EA, Stevens PF, Donoghue MJ (2002) Plant systematics: a phylogenetic approach. 2nd ed. Sunderland, Massachusetts: Sinauer Associates.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

