The IS2 Element Improves Transcription Efficiency of Integration-Deficient Lentiviral Vector Episomes
- PMID: 30227274
- PMCID: PMC6141704
- DOI: 10.1016/j.omtn.2018.08.007
The IS2 Element Improves Transcription Efficiency of Integration-Deficient Lentiviral Vector Episomes
Abstract
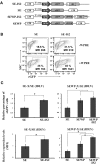
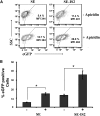
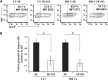
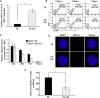
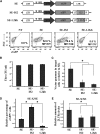
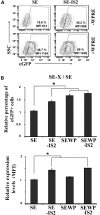
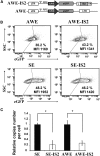
Integration-defective lentiviral vectors (IDLVs) have become an important alternative tool for gene therapy applications and basic research. Unfortunately, IDLVs show lower transgene expression as compared to their integrating counterparts. In this study, we aimed to improve the expression levels of IDLVs by inserting the IS2 element, which harbors SARs and HS4 sequences, into their LTRs (SE-IS2-IDLVs). Contrary to our expectations, the presence of the IS2 element did not abrogate epigenetic silencing by histone deacetylases. In addition, the IS2 element reduced episome levels in IDLV-transduced cells. Interestingly, despite these negative effects, SE-IS2-IDLVs outperformed SE-IDLVs in terms of percentage and expression levels of the transgene in several cell lines, including neurons, neuronal progenitor cells, and induced pluripotent stem cells. We estimated that the IS2 element enhances the transcriptional activity of IDLV LTR circles 6- to 7-fold. The final effect the IS2 element in IDLVs will greatly depend on the target cell and the balance between the negative versus the positive effects of the IS2 element in each cell type. The better performance of SE-IS2-IDLVs was not due to improved stability or differences in the proportions of 1-LTR versus 2-LTR circles but probably to a re-positioning of IS2-episomes into transcriptionally active regions.
Keywords: HS4 insulator; IDLV; gene therapy; lentiviral vector; scaffold or matrix attachment regions.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Improved Functionality of Integration-Deficient Lentiviral Vectors (IDLVs) by the Inclusion of IS2 Protein Docks.Pharmaceutics. 2021 Aug 6;13(8):1217. doi: 10.3390/pharmaceutics13081217. Pharmaceutics. 2021. PMID: 34452178 Free PMC article.
-
A chimeric HS4-SAR insulator (IS2) that prevents silencing and enhances expression of lentiviral vectors in pluripotent stem cells.PLoS One. 2014 Jan 6;9(1):e84268. doi: 10.1371/journal.pone.0084268. eCollection 2014. PLoS One. 2014. PMID: 24400083 Free PMC article.
-
Histone deacetylase inhibition activates transgene expression from integration-defective lentiviral vectors in dividing and non-dividing cells.Hum Gene Ther. 2013 Jan;24(1):78-96. doi: 10.1089/hum.2012.069. Epub 2012 Dec 11. Hum Gene Ther. 2013. PMID: 23140481 Free PMC article.
-
Integration-deficient lentiviral vectors: a slow coming of age.Mol Ther. 2009 Aug;17(8):1316-32. doi: 10.1038/mt.2009.122. Epub 2009 Jun 2. Mol Ther. 2009. PMID: 19491821 Free PMC article. Review.
-
Integrase-defective lentiviral vectors--a stage for nonviral integration machineries.Curr Gene Ther. 2011 Oct;11(5):350-62. doi: 10.2174/156652311797415881. Curr Gene Ther. 2011. PMID: 21745178 Review.
Cited by
-
Genome-edited adult stem cells: Next-generation advanced therapy medicinal products.Stem Cells Transl Med. 2020 Jun;9(6):674-685. doi: 10.1002/sctm.19-0338. Epub 2020 Mar 6. Stem Cells Transl Med. 2020. PMID: 32141715 Free PMC article. Review.
-
CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications.Pharmaceutics. 2021 Oct 9;13(10):1649. doi: 10.3390/pharmaceutics13101649. Pharmaceutics. 2021. PMID: 34683943 Free PMC article. Review.
-
Lentiviral vectors for inducible, transactivator-free advanced therapy medicinal products: Application to CAR-T cells.Mol Ther Nucleic Acids. 2023 Mar 28;32:322-339. doi: 10.1016/j.omtn.2023.03.018. eCollection 2023 Jun 13. Mol Ther Nucleic Acids. 2023. PMID: 37125150 Free PMC article.
-
Characterization of the cHS4 insulator in mouse embryonic stem cells.FEBS Open Bio. 2020 Apr;10(4):644-656. doi: 10.1002/2211-5463.12818. Epub 2020 Mar 19. FEBS Open Bio. 2020. PMID: 32087050 Free PMC article.
-
CRISPR-Based Tools for Fighting Rare Diseases.Life (Basel). 2022 Nov 24;12(12):1968. doi: 10.3390/life12121968. Life (Basel). 2022. PMID: 36556333 Free PMC article. Review.
References
-
- Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. - PubMed
-
- Biffi A., Aubourg P., Cartier N. Gene therapy for leukodystrophies. Hum. Mol. Genet. 2011;20(R1):R42–R53. - PubMed
-
- Biffi A., Montini E., Lorioli L., Cesani M., Fumagalli F., Plati T., Baldoli C., Martino S., Calabria A., Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. - PubMed
-
- Kato S., Kobayashi K., Kobayashi K. Improved transduction efficiency of a lentiviral vector for neuron-specific retrograde gene transfer by optimizing the junction of fusion envelope glycoprotein. J. Neurosci. Methods. 2014;227:151–158. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

