Epigenetic crossroads of the Epstein-Barr virus B-cell relationship
- PMID: 30227386
- PMCID: PMC6263794
- DOI: 10.1016/j.coviro.2018.08.012
Epigenetic crossroads of the Epstein-Barr virus B-cell relationship
Abstract
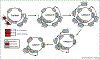
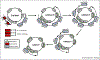
Epstein-Barr virus (EBV) is a gamma-herpesvirus that establishes lifelong infection in the majority of people worldwide. EBV uses epigenetic reprogramming to switch between multiple latency states in order to colonize the memory B-cell compartment and to then periodically undergo lytic reactivation upon plasma cell differentiation. This review focuses on recent advances in the understanding of epigenetic mechanisms that EBV uses to control its lifecycle and to subvert the growth and survival pathways that underly EBV-driven B-cell differentiation versus B-cell growth transformation, a hallmark of the first human tumor virus. These include the formation of viral super enhancers that drive expression of key host dependency factors, evasion of tumor suppressor responses, prevention of plasmablast differentiation, and regulation of the B-cell lytic switch.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures


References
-
- Longnecker R, Kieff E and Cohen JI: Epstein-Barr Virus In Fields Virology, edn 6 Edited by Knipe DMaH PM: Lippincott, Williams and Wilkins; 2013:1898–1959. vol 2.]
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

