Biomolecular Clusters Distribution up to Mega Dalton Region Using MALDI-Quadrupole Ion Trap Mass Spectrometer
- PMID: 30227603
- PMCID: PMC6164314
- DOI: 10.3390/ijms19092789
Biomolecular Clusters Distribution up to Mega Dalton Region Using MALDI-Quadrupole Ion Trap Mass Spectrometer
Abstract
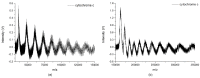
We present the first report on complete cluster distributions of cytochrome c (molecular weight of 12.4 kDa) and bovine serum albumin ((BSA), molecular weight of 66.4 kDa) with mass-to-charge ratio (m/z) reaching 350,000 and 1,400,000, respectively, by matrix-assisted laser desorption/ionization (MALDI). Large cluster distributions of the analytes were measured by our homemade frequency-scanned quadrupole ion trap (QIT) mass spectrometer with a charge detector. To our knowledge, we report the highest m/z clusters of these two biomolecules. The quantitative results indicate that large clusters ions of cytochrome c and BSA follow the power law (r² > 0.99) with cluster size distribution, which provides experimental evidence for the laser ablation studies of MALDI.
Keywords: MALDI; bovine serum albumin (BSA); cytochrome c; large clusters; quantitative distribution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Karas M., Bachmann D., Bahr U., Hillenkamp F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int. J. Mass Spectrom. Ion Process. 1987;78:53–68. doi: 10.1016/0168-1176(87)87041-6. - DOI
-
- Tanaka K., Waki H., Ido Y., Akita S., Yoshida Y., Yoshida T. Protein and polymer analyses up to m/z 100,000 by laser ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1988;2:151–153. doi: 10.1002/rcm.1290020802. - DOI
-
- Taranenko N.I., Potter N.T., Allman S.L., Golovlev V.V., Chen C.H. Detection of trinucleotide expansion in neurodegenerative disease by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Genet. Anal. Biomol. Eng. 1999;15:25–31. doi: 10.1016/S1050-3862(98)00034-5. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

