CDK9: a signaling hub for transcriptional control
- PMID: 30227759
- PMCID: PMC6602564
- DOI: 10.1080/21541264.2018.1523668
CDK9: a signaling hub for transcriptional control
Abstract
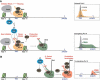
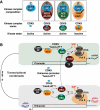
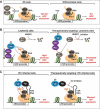
Cyclin-dependent kinase 9 (CDK9) is critical for RNA Polymerase II (Pol II) transcription initiation, elongation, and termination in several key biological processes including development, differentiation, and cell fate responses. A broad range of diseases are characterized by CDK9 malfunction, illustrating its importance in maintaining transcriptional homeostasis in basal- and signal-regulated conditions. Here we provide a historical recount of CDK9 discovery and the current models suggesting CDK9 is a central hub necessary for proper execution of different steps in the transcription cycle. Finally, we discuss the current therapeutic strategies to treat CDK9 malfunction in several disease states. Abbreviations: CDK: Cyclin-dependent kinase; Pol II: RNA Polymerase II; PIC: Pre-initiation Complex; TFIIH: Transcription Factor-II H; snoRNA: small nucleolar RNA; CycT: CyclinT1/T2; P-TEFb: Positive Transcription Elongation Factor Complex; snRNP: small nuclear ribonucleo-protein; HEXIM: Hexamethylene Bis-acetamide-inducible Protein 1/2; LARP7: La-related Protein 7; MePCE: Methylphosphate Capping Enzyme; HIV: human immunodeficiency virus; TAT: trans-activator of transcription; TAR: Trans-activation response element; Hsp70: Heat Shock Protein 70; Hsp90/Cdc37: Hsp90- Hsp90 co-chaperone Cdc37; DSIF: DRB Sensitivity Inducing Factor; NELF: Negative Elongation Factor; CPSF: cleavage and polyadenylation-specific factor; CSTF: cleavage-stimulatory factor; eRNA: enhancer RNA; BRD4: Bromodomain-containing protein 4; JMJD6: Jumonji C-domain-containing protein 6; SEC: Super Elongation Complex; ELL: eleven-nineteen Lys-rich leukemia; ENL: eleven-nineteen leukemia; MLL: mixed lineage leukemia; BEC: BRD4-containing Elongation Complex; SEC-L2/L3: SEC-like complexes; KAP1: Kruppel-associated box-protein 1; KEC: KAP1-7SK Elongation Complex; DRB: Dichloro-1-ß-D-Ribofuranosylbenzimidazole; H2Bub1: H2B mono-ubiquitination; KM: KM05382; PP1: Protein Phosphatase 1; CDK9i: CDK9 inhibitor; SHAPE: Selective 2'-hydroxyl acylation analyzed by primer extension; TE: Typical enhancer; SE : Super enhancer.
Keywords: 7SK; CDK9; Cancer; Disease; Elongation; Enhancer; HIV; P-TEFb; Pausing; RNA polymerase II; Transcription.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
