Genome-Wide Association Study Reveals Genetic Link between Diarrhea-Associated Entamoeba histolytica Infection and Inflammatory Bowel Disease
- PMID: 30228239
- PMCID: PMC6143743
- DOI: 10.1128/mBio.01668-18
Genome-Wide Association Study Reveals Genetic Link between Diarrhea-Associated Entamoeba histolytica Infection and Inflammatory Bowel Disease
Abstract
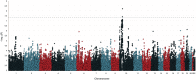
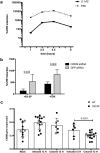
Entamoeba histolytica is the etiologic agent of amebic dysentery, though clinical manifestation of infection is highly variable ranging from subclinical colonization to invasive disease. We hypothesize that host genetics contribute to the variable outcomes of E. histolytica infection; thus, we conducted a genome-wide association study (GWAS) in two independent birth cohorts of Bangladeshi infants monitored for susceptibility to E. histolytica disease in the first year of life. Children with at least one diarrheal episode positive for E. histolytica (cases) were compared to children with no detectable E. histolytica infection in the same time frame (controls). Meta-analyses under a fixed-effect inverse variance weighting model identified multiple variants in a region of chromosome 10 containing loci associated with symptomatic E. histolytica infection. An intergenic insertion between CREM and CCNY (rs58000832) achieved genome-wide significance (P value from meta-analysis [Pmeta] = 6.05 × 10-9), and each additional risk allele of rs58000832 conferred 2.42 increased odds of a diarrhea-associated E. histolytica infection. The most strongly associated single nucleotide polymorphism (SNP) within a gene was in an intron of CREM (rs58468612; Pmeta = 8.94 × 10-8), which has been implicated as a susceptibility locus for inflammatory bowel disease (IBD). Gene expression resources suggest associated loci are related to the lower expression of CREM Increased CREM expression is also observed in early E. histolytica infection. Further, CREM-/- mice were more susceptible to E. histolytica amebic colitis. These genetic associations reinforce the pathological similarities observed in gut inflammation between E. histolytica infection and IBD.IMPORTANCE Diarrhea is the second leading cause of death for children globally, causing 760,000 deaths each year in children less than 5 years old. Amebic dysentery contributes significantly to this burden, especially in developing countries. The identification of host factors that control or enable enteric pathogens has the potential to transform our understanding of disease predisposition, outcomes, and treatments. Our discovery of the transcriptional regulator cAMP-responsive element modulator (CREM) as a genetic modifier of susceptibility to amebic disease has implications for understanding the pathogenesis of other diarrheal infections. Further, emerging evidence for CREM in IBD susceptibility suggests that CREM is a critical regulator of enteric inflammation and may have broad therapeutic potential as a drug target across intestinal inflammatory diseases.
Keywords: diarrhea; epidemiology; genomics; infectious disease; protozoa; public health.
Copyright © 2018 Wojcik et al.
Figures


References
-
- United Nations. 2015. The Millennium Development Goals Report, p 1–15. United Nations, New York, NY.
-
- Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE. 2015. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 385:430–440. doi: 10.1016/S0140-6736(14)61698-6. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

