Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation
- PMID: 30228257
- PMCID: PMC6143631
- DOI: 10.1038/s41467-018-06237-7
Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation
Abstract
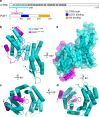
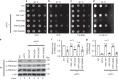
The evolutionarily conserved multifunctional polymerase-associated factor 1 (Paf1) complex (Paf1C), which is composed of at least five subunits (Paf1, Leo1, Ctr9, Cdc73, and Rtf1), plays vital roles in gene regulation and has connections to development and human diseases. Here, we report two structures of each of the human and yeast Ctr9/Paf1 subcomplexes, which assemble into heterodimers with very similar conformations, revealing an interface between the tetratricopeptide repeat module in Ctr9 and Paf1. The structure of the Ctr9/Paf1 subcomplex may provide mechanistic explanations for disease-associated mutations in human PAF1 and CTR9. Our study reveals that the formation of the Ctr9/Paf1 heterodimer is required for the assembly of yeast Paf1C, and is essential for yeast viability. In addition, disruption of the interaction between Paf1 and Ctr9 greatly affects the level of histone H3 methylation in vivo. Collectively, our results shed light on Paf1C assembly and functional regulation.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Biochemical insights into Paf1 complex-induced stimulation of Rad6/Bre1-mediated H2B monoubiquitination.Proc Natl Acad Sci U S A. 2021 Aug 17;118(33):e2025291118. doi: 10.1073/pnas.2025291118. Proc Natl Acad Sci U S A. 2021. PMID: 34385316 Free PMC article.
-
Crystal Structure of the Core Module of the Yeast Paf1 Complex.J Mol Biol. 2022 Jan 30;434(2):167369. doi: 10.1016/j.jmb.2021.167369. Epub 2021 Nov 28. J Mol Biol. 2022. PMID: 34852272
-
Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):9998-10003. doi: 10.1073/pnas.1812256115. Epub 2018 Sep 17. Proc Natl Acad Sci U S A. 2018. PMID: 30224485 Free PMC article.
-
The Paf1 complex: platform or player in RNA polymerase II transcription?Biochim Biophys Acta. 2010 May-Jun;1799(5-6):379-88. doi: 10.1016/j.bbagrm.2010.01.001. Epub 2010 Jan 12. Biochim Biophys Acta. 2010. PMID: 20060942 Free PMC article. Review.
-
Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation.Trends Biochem Sci. 2017 Oct;42(10):788-798. doi: 10.1016/j.tibs.2017.08.003. Epub 2017 Sep 1. Trends Biochem Sci. 2017. PMID: 28870425 Free PMC article. Review.
Cited by
-
The Paf1 Complex: A Keystone of Nuclear Regulation Operating at the Interface of Transcription and Chromatin.J Mol Biol. 2021 Jul 9;433(14):166979. doi: 10.1016/j.jmb.2021.166979. Epub 2021 Apr 1. J Mol Biol. 2021. PMID: 33811920 Free PMC article. Review.
-
The Paf1 complex transcriptionally regulates the mitochondrial-anchored protein Atg32 leading to activation of mitophagy.Autophagy. 2020 Aug;16(8):1366-1379. doi: 10.1080/15548627.2019.1668228. Epub 2019 Sep 19. Autophagy. 2020. PMID: 31525119 Free PMC article.
-
High-resolution Sequencing Reveals that the Paf1 Complex May be a Conserved Transcription Elongation Factor for Eukaryotic RNA Polymerase I.J Mol Biol. 2025 Sep 1;437(17):169220. doi: 10.1016/j.jmb.2025.169220. Epub 2025 May 19. J Mol Biol. 2025. PMID: 40398673
-
Biochemical insights into Paf1 complex-induced stimulation of Rad6/Bre1-mediated H2B monoubiquitination.Proc Natl Acad Sci U S A. 2021 Aug 17;118(33):e2025291118. doi: 10.1073/pnas.2025291118. Proc Natl Acad Sci U S A. 2021. PMID: 34385316 Free PMC article.
-
Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation.Microorganisms. 2021 Aug 30;9(9):1837. doi: 10.3390/microorganisms9091837. Microorganisms. 2021. PMID: 34576732 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

