Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum
- PMID: 30228375
- PMCID: PMC6143587
- DOI: 10.1038/s41598-018-32080-3
Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum
Abstract
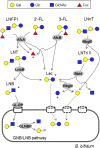
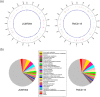
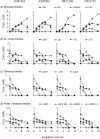
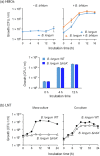
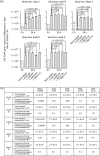
Gut microbiota of breast-fed infants are generally rich in bifidobacteria. Recent studies show that infant gut-associated bifidobacteria can assimilate human milk oligosaccharides (HMOs) specifically among the gut microbes. Nonetheless, little is known about how bifidobacterial-rich communities are shaped in the gut. Interestingly, HMOs assimilation ability is not related to the dominance of each species. Bifidobacterium longum susbp. longum and Bifidobacterium breve are commonly found as the dominant species in infant stools; however, they show limited HMOs assimilation ability in vitro. In contrast, avid in vitro HMOs consumers, Bifidobacterium bifidum and Bifidobacterium longum subsp. infantis, are less abundant in infant stools. In this study, we observed altruistic behaviour by B. bifidum when incubated in HMOs-containing faecal cultures. Four B. bifidum strains, all of which contained complete sets of HMO-degrading genes, commonly left HMOs degradants unconsumed during in vitro growth. These strains stimulated the growth of other Bifidobacterium species when added to faecal cultures supplemented with HMOs, thereby increasing the prevalence of bifidobacteria in faecal communities. Enhanced HMOs consumption by B. bifidum-supplemented cultures was also observed. We also determined the complete genome sequences of B. bifidum strains JCM7004 and TMC3115. Our results suggest B. bifidum-mediated cross-feeding of HMOs degradants within bifidobacterial communities.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

