Analysis of Transcription Factor-Related Regulatory Networks Based on Bioinformatics Analysis and Validation in Hepatocellular Carcinoma
- PMID: 30228980
- PMCID: PMC6136478
- DOI: 10.1155/2018/1431396
Analysis of Transcription Factor-Related Regulatory Networks Based on Bioinformatics Analysis and Validation in Hepatocellular Carcinoma
Abstract
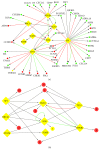
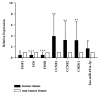
Hepatocellular carcinoma (HCC) accounts for a significant proportion of liver cancer, which has become the second most common cause of cancer-related mortality worldwide. To investigate the potential mechanisms of invasion and progression of HCC, bioinformatics analysis and validation by qRT-PCR were performed. We found 237 differentially expressed genes (DEGs) including EGR1, FOS, and FOSB, which were three cancer-related transcription factors. Subsequently, we constructed TF-gene network and miRNA-TF-mRNA network based on data obtained from mRNA and miRNA expression profiles for analysis of HCC. We found that 42 key genes from the TF-gene network including EGR1, FOS, and FOSB were most enriched in the p53 signaling pathway. The qRT-PCR data confirmed that mRNA levels of EGR1, FOS, and FOSB all were decreased in HCC tissues. In addition, we confirmed that the mRNA levels of CCNB1, CCNB2, and CHEK1, three key markers of the p53 signaling pathway, were all increased in HCC tissues by bioinformatics analysis and qRT-PCR validation. Therefore, we speculated that miR-181a-5p, which was upregulated in HCC tissues, could regulate FOS and EGR1 to promote the invasion and progression of HCC by p53 signaling pathway. Overall, the study provides support for the possible mechanisms of progression in HCC.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

