Quantitative Analysis of the Brain Ubiquitylome in Alzheimer's Disease
- PMID: 30230243
- PMCID: PMC6283072
- DOI: 10.1002/pmic.201800108
Quantitative Analysis of the Brain Ubiquitylome in Alzheimer's Disease
Abstract
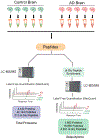
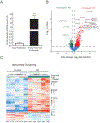
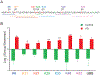
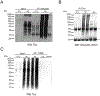
Several neurodegenerative diseases including Alzheimer's Disease (AD) are characterized by ubiquitin-positive pathological protein aggregates. Here, an immunoaffinity approach is utilized to enrich ubiquitylated isopeptides after trypsin digestion from five AD and five age-matched control postmortem brain tissues. Label-free MS-based proteomic analysis identifies 4291 unique ubiquitylation sites mapping to 1682 unique proteins. Differential enrichment analysis shows that over 800 ubiquitylation sites are significantly altered between AD and control cases. Of these, ≈80% are increased in AD, including seven poly ubiquitin linkages, which is consistent with proteolytic stress and high burden of ubiquitylated pathological aggregates in AD. The microtubule associated protein Tau, the core component of neurofibrillary tangles, has the highest number of increased sites of ubiquitylation per any protein in AD. Tau poly ubiquitylation from AD brain homogenates is confirmed by reciprocal co-immunoprecipitation and by affinity capture using tandem ubiquitin binding entities. Co-modified peptides, with both ubiquitylation and phosphorylation sites, are also enriched in AD. Notably, many of the co-modified peptides mapped to Tau within KXGS motifs in the microtubule binding region suggesting that crosstalk between phosphorylation and ubiquitylation occurs on Tau in AD. Overall, these findings highlight the utility of MS to map ubiquitylated substrates in human brain and provides insight into mechanisms underlying pathological protein posttranslational modification in AD.
Keywords: mass spectrometry; neurodegeneration; post-translational modifications; protein aggregation; proteomics; ubiquitylation.
© 2018 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures



References
-
- Bence NF, Sampat RM, Kopito RR, Science 2001, 292, 1552. - PubMed
-
- Gilberto S, Peter M, The Journal of cell biology 2017, 216, 2259; - PMC - PubMed
- Grillari J, Grillari Voglauer R, Jansen-Durr P, Advances in experimental medicine and biology 2010, 694, 172; - PubMed
- Muratani M, Tansey WP, Nature reviews Molecular cell biology 2003, 4, 192; - PubMed
- Ciechanover A, The EMBO journal 1998, 17, 7151; - PMC - PubMed
- Hershko A, Ciechanover A, Annual Reviews 4139 El Camino Way, PO Box 10139, Palo Alto, CA 94303–0139, USA, 1998.
-
- Scheffner M, Nuber U, Huibregtse JM, Nature 1995, 373, 81. - PubMed
-
- Kerscher O, Felberbaum R, Hochstrasser M, Annu. Rev. Cell Dev. Biol 2006, 22, 159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- RF1 AG057471/AG/NIA NIH HHS/United States
- RF1AG057471/AG/NIA NIH HHS/United States
- R01 AG053960/AG/NIA NIH HHS/United States
- ALZ/Alzheimer's Association/United States
- 5R01AG053960/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- R21AG054206/AG/NIA NIH HHS/United States
- T32 NS007480/NS/NINDS NIH HHS/United States
- U01AG046161-02/AG/NIA NIH HHS/United States
- T32 NS00748018/NS/NINDS NIH HHS/United States
- RF1 AG057470/AG/NIA NIH HHS/United States
- P30 NS055077/NS/NINDS NIH HHS/United States
- P50AG025688/AG/NIA NIH HHS/United States
- P30NS055077/NS/NINDS NIH HHS/United States
- R21 AG054206/AG/NIA NIH HHS/United States
- U01 AG046161/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

