Tip60 complex promotes expression of a differentiation factor to regulate germline differentiation in female Drosophila
- PMID: 30230973
- PMCID: PMC6329907
- DOI: 10.1091/mbc.E18-06-0385
Tip60 complex promotes expression of a differentiation factor to regulate germline differentiation in female Drosophila
Abstract
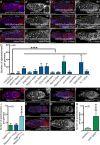
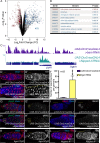
Germline stem cells (GSCs) self-renew and differentiate to sustain a continuous production of gametes. In the female Drosophila germ line, two differentiation factors, bag of marbles ( bam) and benign gonial cell neoplasm ( bgcn), work in concert in the stem cell daughter to promote the generation of eggs. In GSCs, bam transcription is repressed by signaling from the niche and is activated in stem cell daughters. In contrast, bgcn is transcribed in both the GSCs and stem cell daughters, but little is known about how bgcn is transcriptionally modulated. Here we find that the conserved protein Nipped-A acts through the Tat interactive protein 60-kDa (Tip60) histone acetyl transferase complex in the germ line to promote GSC daughter differentiation. We find that Nipped-A is required for efficient exit from the gap phase 2 (G2) of cell cycle of the GSC daughter and for expression of a differentiation factor, bgcn. Loss of Nipped-A results in accumulation of GSC daughters . Forced expression of bgcn in Nipped-A germline-depleted ovaries rescues this differentiation defect. Together, our results indicate that Tip60 complex coordinates cell cycle progression and expression of bgcn to help drive GSC daughters toward a differentiation program.
Figures



References
-
- Abdu U, Brodsky M, Schüpbach T. (2002). Activation of a meiotic checkpoint during Drosophila oogenesis regulates the translation of Gurken through Chk2/Mnk. Curr Biol , 1645–1651. - PubMed
-
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. (2005). c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol , 303–310. - PubMed
-
- Avvakumov N, Côté J. (2007a). Functions of myst family histone acetyltransferases and their link to disease. Subcell Biochem , 295–317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

