Stress-Induced Mucus Secretion and Its Composition by a Combination of Proteomics and Metabolomics of the Jellyfish Aurelia coerulea
- PMID: 30231483
- PMCID: PMC6165293
- DOI: 10.3390/md16090341
Stress-Induced Mucus Secretion and Its Composition by a Combination of Proteomics and Metabolomics of the Jellyfish Aurelia coerulea
Abstract
Background: Jellyfish respond quickly to external stress that stimulates mucus secretion as a defense. Neither the composition of secreted mucus nor the process of secretion are well understood.
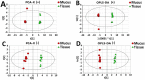
Methods: Aurelia coerulea jellyfish were stimulated by removing them from environmental seawater. Secreted mucus and tissue samples were then collected within 60 min, and analyzed by a combination of proteomics and metabolomics using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS/MS), respectively.
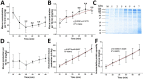
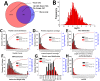
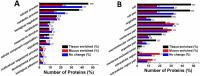
Results: Two phases of sample collection displayed a quick decrease in volume, followed by a gradual increase. A total of 2421 and 1208 proteins were identified in tissue homogenate and secreted mucus, respectively. Gene Ontology (GO) analysis showed that the mucus-enriched proteins are mainly located in extracellular or membrane-associated regions, while the tissue-enriched proteins are distributed throughout intracellular compartments. Tryptamine, among 16 different metabolites, increased with the largest-fold change value of 7.8 in mucus, which is consistent with its involvement in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway 'tryptophan metabolism'. We identified 11 metalloproteinases, four serpins, three superoxide dismutases and three complements, and their presence was speculated to be related to self-protective defense.
Conclusions: Our results provide a composition profile of proteins and metabolites in stress-induced mucus and tissue homogenate of A. coerulea. This provides insight for the ongoing endeavors to discover novel bioactive compounds. The large increase of tryptamine in mucus may indicate a strong stress response when jellyfish were taken out of seawater and the active self-protective components such as enzymes, serpins and complements potentially play a key role in innate immunity of jellyfish.
Keywords: Aurelia coerulea; jellyfish; metabolomics; mucus; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Comparative proteomic analysis of Aurelia coerulea for its locomotion system molecular structure-function inference.J Proteomics. 2019 Oct 30;209:103509. doi: 10.1016/j.jprot.2019.103509. Epub 2019 Aug 31. J Proteomics. 2019. PMID: 31479797
-
Combined effects of ocean acidification and temperature on planula larvae of the moon jellyfish Aurelia coerulea.Mar Environ Res. 2018 Aug;139:144-150. doi: 10.1016/j.marenvres.2018.05.015. Epub 2018 May 16. Mar Environ Res. 2018. PMID: 29789135
-
Jellyfish Bioprospecting in the Mediterranean Sea: Antioxidant and Lysozyme-Like Activities from Aurelia coerulea (Cnidaria, Scyphozoa) Extracts.Mar Drugs. 2021 Oct 31;19(11):619. doi: 10.3390/md19110619. Mar Drugs. 2021. PMID: 34822490 Free PMC article.
-
UPLC-MS(E) application in disease biomarker discovery: the discoveries in proteomics to metabolomics.Chem Biol Interact. 2014 May 25;215:7-16. doi: 10.1016/j.cbi.2014.02.014. Epub 2014 Mar 12. Chem Biol Interact. 2014. PMID: 24631021 Review.
-
[Advances in liquid chromatography-mass spectrometry analysis of several important secondary metabolites in plant metabolomics].Sheng Wu Gong Cheng Xue Bao. 2022 Oct 25;38(10):3674-3681. doi: 10.13345/j.cjb.220488. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 36305402 Review. Chinese.
Cited by
-
Chemical Defenses in Medusozoa.Mar Drugs. 2025 May 28;23(6):229. doi: 10.3390/md23060229. Mar Drugs. 2025. PMID: 40559638 Free PMC article. Review.
-
Jellyfish from Fisheries By-Catches as a Sustainable Source of High-Value Compounds with Biotechnological Applications.Mar Drugs. 2022 Apr 14;20(4):266. doi: 10.3390/md20040266. Mar Drugs. 2022. PMID: 35447939 Free PMC article. Review.
-
Comparative mucomic analysis of three functionally distinct Cornu aspersum Secretions.Nat Commun. 2023 Sep 2;14(1):5361. doi: 10.1038/s41467-023-41094-z. Nat Commun. 2023. PMID: 37660066 Free PMC article.
-
Experimental mining plumes and ocean warming trigger stress in a deep pelagic jellyfish.Nat Commun. 2023 Nov 21;14(1):7352. doi: 10.1038/s41467-023-43023-6. Nat Commun. 2023. PMID: 37990021 Free PMC article.
-
Marine Bioprospecting: Enzymes and Stress Proteins from the Sea Anemones Anthopleura dowii and Lebrunia neglecta.Mar Drugs. 2023 Dec 23;22(1):12. doi: 10.3390/md22010012. Mar Drugs. 2023. PMID: 38248637 Free PMC article.
References
-
- Zhang L., He Q., Wang Q., Zhang B., Wang B., Xu F., Wang T., Xiao L., Zhang L. Intracellular Ca2+ overload induced by extracellular Ca2+ entry plays an important role in acute heart dysfunction by tentacle extract from the jellyfish Cyanea capillata. Cardiovasc. Toxicol. 2014;14:260–274. doi: 10.1007/s12012-014-9250-6. - DOI - PubMed
-
- Lazcanopérez F., Arellano R.O., Garay E., Arreguínespinosa R., Sánchezrodríguez J. Electrophysiological activity of a neurotoxic fraction from the venom of box jellyfish Carybdea marsupialis. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017;191:177–182. doi: 10.1016/j.cbpc.2016.10.010. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

