Bacterial endosymbiont Cardinium cSfur genome sequence provides insights for understanding the symbiotic relationship in Sogatella furcifera host
- PMID: 30231855
- PMCID: PMC6147030
- DOI: 10.1186/s12864-018-5078-y
Bacterial endosymbiont Cardinium cSfur genome sequence provides insights for understanding the symbiotic relationship in Sogatella furcifera host
Abstract
Background: Sogatella furcifera is a migratory pest that damages rice plants and causes severe economic losses. Due to its ability to annually migrate long distances, S. furcifera has emerged as a major pest of rice in several Asian countries. Symbiotic relationships of inherited bacteria with terrestrial arthropods have significant implications. The genus Cardinium is present in many types of arthropods, where it influences some host characteristics. We present a report of a newly identified strain of the bacterial endosymbiont Cardinium cSfur in S. furcifera.
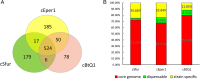
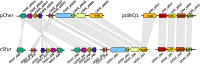
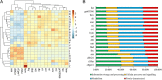
Result: From the whole genome of S. furcifera previously sequenced by our laboratory, we assembled the whole genome sequence of Cardinium cSfur. The sequence comprised 1,103,593 bp with a GC content of 39.2%. The phylogenetic tree of the Bacteroides phylum to which Cardinium cSfur belongs suggests that Cardinium cSfur is closely related to the other strains (Cardinium cBtQ1 and cEper1) that are members of the Amoebophilaceae family. Genome comparison between the host-dependent endosymbiont including Cardinium cSfur and free-living bacteria revealed that the endosymbiont has a smaller genome size and lower GC content, and has lost some genes related to metabolism because of its special environment, which is similar to the genome pattern observed in other insect symbionts. Cardinium cSfur has limited metabolic capability, which makes it less contributive to metabolic and biosynthetic processes in its host. From our findings, we inferred that, to compensate for its limited metabolic capability, Cardinium cSfur harbors a relatively high proportion of transport proteins, which might act as the hub between it and its host. With its acquisition of the whole operon related to biotin synthesis and glycolysis related genes through HGT event, Cardinium cSfur seems to be undergoing changes while establishing a symbiotic relationship with its host.
Conclusion: A novel bacterial endosymbiont strain (Cardinium cSfur) has been discovered. A genomic analysis of the endosymbiont in S. furcifera suggests that its genome has undergone certain changes to facilitate its settlement in the host. The envisaged potential reproduction manipulative ability of the new endosymbiont strain in its S. furcifera host has vital implications in designing eco-friendly approaches to combat the insect pest.
Keywords: Bacterial endosymbionts; Cardinium; Genomic analysis; Sogatella furcifera.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





Similar articles
-
Endosymbionts Reduce Microbiome Diversity and Modify Host Metabolism and Fecundity in the Planthopper Sogatella furcifera.mSystems. 2022 Apr 26;7(2):e0151621. doi: 10.1128/msystems.01516-21. Epub 2022 Mar 30. mSystems. 2022. PMID: 35353007 Free PMC article.
-
The genome of Cardinium cBtQ1 provides insights into genome reduction, symbiont motility, and its settlement in Bemisia tabaci.Genome Biol Evol. 2014 Apr;6(4):1013-30. doi: 10.1093/gbe/evu077. Genome Biol Evol. 2014. PMID: 24723729 Free PMC article.
-
Expression of cytoplasmic incompatibility and host fitness effects in field populations of Sogatella furcifera infected with Cardinium.J Econ Entomol. 2012 Dec;105(6):2161-6. doi: 10.1603/ec12268. J Econ Entomol. 2012. PMID: 23356082
-
Bacterial reproductive manipulators in rice planthoppers.Arch Insect Biochem Physiol. 2019 Jun;101(2):e21548. doi: 10.1002/arch.21548. Epub 2019 Mar 25. Arch Insect Biochem Physiol. 2019. PMID: 30912174 Review.
-
Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10255-61. doi: 10.1073/pnas.1423305112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26039986 Free PMC article. Review.
Cited by
-
Endosymbionts Reduce Microbiome Diversity and Modify Host Metabolism and Fecundity in the Planthopper Sogatella furcifera.mSystems. 2022 Apr 26;7(2):e0151621. doi: 10.1128/msystems.01516-21. Epub 2022 Mar 30. mSystems. 2022. PMID: 35353007 Free PMC article.
-
Development of a multi-locus sequence typing system helps reveal the evolution of Cardinium hertigii, a reproductive manipulator symbiont of insects.BMC Microbiol. 2019 Nov 27;19(1):266. doi: 10.1186/s12866-019-1638-9. BMC Microbiol. 2019. PMID: 31775631 Free PMC article.
-
A Comparative Analyses of the Complete Mitochondrial Genomes of Fungal Endosymbionts in Sogatella furcifera, White-Backed Planthoppers.Int J Genomics. 2021 Jun 8;2021:6652508. doi: 10.1155/2021/6652508. eCollection 2021. Int J Genomics. 2021. PMID: 34212028 Free PMC article.
-
One to host them all: genomics of the diverse bacterial endosymbionts of the spider Oedothorax gibbosus.Microb Genom. 2023 Feb;9(2):mgen000943. doi: 10.1099/mgen.0.000943. Microb Genom. 2023. PMID: 36757767 Free PMC article.
-
Both symbionts and environmental factors contribute to shape the microbiota in a pest insect, Sogatella furcifera.Front Microbiol. 2024 Jan 24;14:1336345. doi: 10.3389/fmicb.2023.1336345. eCollection 2023. Front Microbiol. 2024. PMID: 38348307 Free PMC article.
References
-
- Zhou WW, Liang QM, Xu Y, Gurr GM, Bao YY, Zhou XP, Zhang CX, Cheng J, Zhu ZR. Genomic Insights into the Glutathione S-Transferase Gene Family of Two Rice Planthoppers, Nilaparvata lugens (Stal) and Sogatella furcifera (Horvath) (Hemiptera: Delphacidae) PLoS One. 2013;8(2):e56604. doi: 10.1371/journal.pone.0056604. - DOI - PMC - PubMed
-
- Ramesh K, Padmavathi G, Deen R, Pandey MK, Lakshmi VJ, Bentur J. Whitebacked planthopper Sogatella furcifera (Horváth)(Homoptera: Delphacidae) resistance in rice variety Sinna Sivappu. Euphytica. 2014;200(1):139–148. doi: 10.1007/s10681-014-1175-4. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

