The Adaptor Protein CARD9 Protects against Colon Cancer by Restricting Mycobiota-Mediated Expansion of Myeloid-Derived Suppressor Cells
- PMID: 30231984
- PMCID: PMC6880241
- DOI: 10.1016/j.immuni.2018.08.018
The Adaptor Protein CARD9 Protects against Colon Cancer by Restricting Mycobiota-Mediated Expansion of Myeloid-Derived Suppressor Cells
Abstract
The adaptor protein CARD9 links detection of fungi by surface receptors to the activation of the NF-κB pathway. Mice deficient in CARD9 exhibit dysbiosis and are more susceptible to colitis. Here we examined the impact of Card9 deficiency in the development of colitis-associated colon cancer (CAC). Treatment of Card9-/- mice with AOM-DSS resulted in increased tumor loads as compared to WT mice and in the accumulation of myeloid-derived suppressor cells (MDSCs) in tumor tissue. The impaired fungicidal functions of Card9-/- macrophages led to increased fungal loads and variation in the overall composition of the intestinal mycobiota, with a notable increase in C. tropicalis. Bone marrow cells incubated with C. tropicalis exhibited MDSC features and suppressive functions. Fluconazole treatment suppressed CAC in Card9-/- mice and was associated with decreased MDSC accumulation. The frequency of MDSCs in tumor tissues of colon cancer patients correlated positively with fungal burden, pointing to the relevance of this regulatory axis in human disease.
Keywords: Card9; MDSCs; colon cancer; mycobiota.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



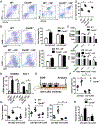

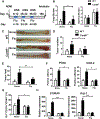
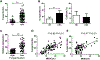
Comment in
-
Fungi Enter the Stage of Colon Carcinogenesis.Immunity. 2018 Sep 18;49(3):384-386. doi: 10.1016/j.immuni.2018.09.002. Immunity. 2018. PMID: 30231977
References
-
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, and Bray F (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66, 683–691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

