A New Tool to Reveal Bacterial Signaling Mechanisms in Antibiotic Treatment and Resistance
- PMID: 30232125
- PMCID: PMC6283303
- DOI: 10.1074/mcp.RA118.000880
A New Tool to Reveal Bacterial Signaling Mechanisms in Antibiotic Treatment and Resistance
Abstract
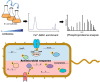
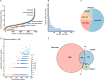
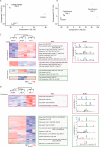
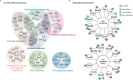
The rapid emergence of antimicrobial resistance is a major threat to human health. Antibiotics modulate a wide range of biological processes in bacteria and as such, the study of bacterial cellular signaling could aid the development of urgently needed new antibiotic agents. Due to the advances in bacterial phosphoproteomics, such a systemwide analysis of bacterial signaling in response to antibiotics has recently become feasible. Here we present a dynamic view of differential protein phosphorylation upon antibiotic treatment and antibiotic resistance. Most strikingly, differential phosphorylation was observed on highly conserved residues of resistance regulating transcription factors, implying a previously unanticipated role of phosphorylation mediated regulation. Using the comprehensive phosphoproteomics data presented here as a resource, future research can now focus on deciphering the precise signaling mechanisms contributing to resistance, eventually leading to alternative strategies to combat antimicrobial resistance.
Keywords: Bacteria; Microbiology; Pathogens; Phosphorylation; Quantification; antimicrobial resistance.
© 2018 Lin et al.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- O'Neill J. (2016) Tackling drug-resistant infections globally: Final report and recommendations https://amr-review.org/sites/default/files/160525_Final%20paper_with%20c...
-
- Hoffman L. R., D'Argenio D. A., MacCoss M. J., Zhang Z., Jones R. A., and Miller S. I. (2005) Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

