Protein palmitoylation and cancer
- PMID: 30232163
- PMCID: PMC6172454
- DOI: 10.15252/embr.201846666
Protein palmitoylation and cancer
Abstract
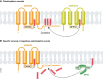
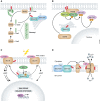
Protein S-palmitoylation is a reversible post-translational modification that alters the localization, stability, and function of hundreds of proteins in the cell. S-palmitoylation is essential for the function of both oncogenes (e.g., NRAS and EGFR) and tumor suppressors (e.g., SCRIB, melanocortin 1 receptor). In mammalian cells, the thioesterification of palmitate to internal cysteine residues is catalyzed by 23 Asp-His-His-Cys (DHHC)-family palmitoyl S-acyltransferases while the removal of palmitate is catalyzed by serine hydrolases, including acyl-protein thioesterases (APTs). These enzymes modulate the function of important oncogenes and tumor suppressors and often display altered expression patterns in cancer. Targeting S-palmitoylation or the enzymes responsible for palmitoylation dynamics may therefore represent a candidate therapeutic strategy for certain cancers.
Keywords: S‐palmitoylation; lipid; lipidation; post‐translational modification; tumor.
© 2018 The Authors.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

