NSs Protein of Sandfly Fever Sicilian Phlebovirus Counteracts Interferon (IFN) Induction by Masking the DNA-Binding Domain of IFN Regulatory Factor 3
- PMID: 30232186
- PMCID: PMC6232482
- DOI: 10.1128/JVI.01202-18
NSs Protein of Sandfly Fever Sicilian Phlebovirus Counteracts Interferon (IFN) Induction by Masking the DNA-Binding Domain of IFN Regulatory Factor 3
Abstract
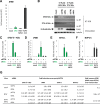
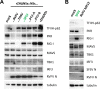
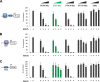
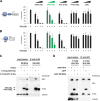
Sandfly fever Sicilian virus (SFSV) is one of the most widespread and frequently identified members of the genus Phlebovirus (order Bunyavirales, family Phenuiviridae) infecting humans. Being transmitted by Phlebotomus sandflies, SFSV causes a self-limiting, acute, often incapacitating febrile disease ("sandfly fever," "Pappataci fever," or "dog disease") that has been known since at least the beginning of the 20th century. We show that, similarly to other pathogenic phleboviruses, SFSV suppresses the induction of the antiviral type I interferon (IFN) system in an NSs-dependent manner. SFSV NSs interfered with the TBK1-interferon regulatory factor 3 (IRF3) branch of the RIG-I signaling pathway but not with NF-κB activation. Consistently, we identified IRF3 as a host interactor of SFSV NSs. In contrast to IRF3, neither the IFN master regulator IRF7 nor any of the related transcription factors IRF2, IRF5, and IRF9 were bound by SFSV NSs. In spite of this specificity for IRF3, NSs did not inhibit its phosphorylation, dimerization, or nuclear accumulation, and the interaction was independent of the IRF3 activation or multimerization state. In further studies, we identified the DNA-binding domain of IRF3 (amino acids 1 to 113) as sufficient for NSs binding and found that SFSV NSs prevented the association of activated IRF3 with the IFN-β promoter. Thus, unlike highly virulent phleboviruses, which either destroy antiviral host factors or sequester whole signaling chains into inactive aggregates, SFSV modulates type I IFN induction by directly masking the DNA-binding domain of IRF3.IMPORTANCE Phleboviruses are receiving increased attention due to the constant discovery of new species and the ongoing spread of long-known members of the genus. Outbreaks of sandfly fever were reported in the 19th century, during World War I, and during World War II. Currently, SFSV is recognized as one of the most widespread phleboviruses, exhibiting high seroprevalence rates in humans and domestic animals and causing a self-limiting but incapacitating disease predominantly in immunologically naive troops and travelers. We show how the nonstructural NSs protein of SFSV counteracts the upregulation of the antiviral interferon (IFN) system. SFSV NSs specifically inhibits promoter binding by IFN transcription factor 3 (IRF3), a molecular strategy which is unique among phleboviruses and, to our knowledge, among human pathogenic RNA viruses in general. This IRF3-specific and stoichiometric mechanism, greatly distinct from the ones exhibited by the highly virulent phleboviruses, correlates with the intermediate level of pathogenicity of SFSV.
Keywords: DNA-binding domain; IRF3; NSs; interferon beta promoter; interferon induction; sandfly fever Sicilian virus.
Copyright © 2018 Wuerth et al.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

