Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome
- PMID: 30232199
- PMCID: PMC6169887
- DOI: 10.1101/gr.236000.118
Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome
Abstract
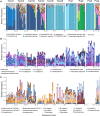
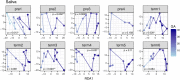
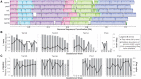
Recent studies suggest that the microbiome has an impact on gestational health and outcome. However, characterization of the pregnancy-associated microbiome has largely relied on 16S rRNA gene amplicon-based surveys. Here, we describe an assembly-driven, metagenomics-based, longitudinal study of the vaginal, gut, and oral microbiomes in 292 samples from 10 subjects sampled every three weeks throughout pregnancy. Nonhuman sequences in the amount of 1.53 Gb were assembled into scaffolds, and functional genes were predicted for gene- and pathway-based analyses. Vaginal assemblies were binned into 97 draft quality genomes. Redundancy analysis (RDA) of microbial community composition at all three body sites revealed gestational age to be a significant source of variation in patterns of gene abundance. In addition, health complications were associated with variation in community functional gene composition in the mouth and gut. The diversity of Lactobacillus iners-dominated communities in the vagina, unlike most other vaginal community types, significantly increased with gestational age. The genomes of co-occurring Gardnerella vaginalis strains with predicted distinct functions were recovered in samples from two subjects. In seven subjects, gut samples contained strains of the same Lactobacillus species that dominated the vaginal community of that same subject and not other Lactobacillus species; however, these within-host strains were divergent. CRISPR spacer analysis suggested shared phage and plasmid populations across body sites and individuals. This work underscores the dynamic behavior of the microbiome during pregnancy and suggests the potential importance of understanding the sources of this behavior for fetal development and gestational outcome.
© 2018 Goltsman et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, et al. 2012. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol 194: 3922–3937. - PMC - PubMed
-
- Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320: 1047–1050. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiol 151: 2551–2561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
