Cell-based RNAi screening and high-content analysis in primary calvarian osteoblasts applied to identification of osteoblast differentiation regulators
- PMID: 30232406
- PMCID: PMC6145911
- DOI: 10.1038/s41598-018-32364-8
Cell-based RNAi screening and high-content analysis in primary calvarian osteoblasts applied to identification of osteoblast differentiation regulators
Abstract
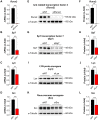
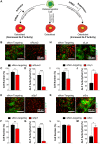
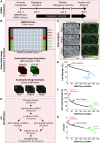
Osteoblasts are responsible for the maintenance of bone homeostasis. Deregulation of their differentiation is etiologically linked to several bone disorders, making this process an important target for therapeutic intervention. Systemic identification of osteoblast regulators has been hampered by the unavailability of physiologically relevant in vitro systems suitable for efficient RNAi and for differentiation read-outs compatible with fluorescent microscopy-based high-content analysis (HCA). Here, we report a new method for identification of osteoblast differentiation regulators by combining siRNA transfection in physiologically relevant cells with high-throughput screening (HTS). Primary mouse calvarial osteoblasts were seeded in 384-well format and reverse transfected with siRNAs and their cell number and differentiation was assayed by HCA. Automated image acquisition allowed high-throughput analyses and classification of single cell features. The physiological relevance, reproducibility, and sensitivity of the method were validated using known regulators of osteoblast differentiation. The application of HCA to siRNAs against expression of 320 genes led to the identification of five potential suppressors and 60 activators of early osteoblast differentiation. The described method and the associated analysis pipeline are not restricted to RNAi-based screening, but can be adapted to large-scale drug HTS or to small-scale targeted experiments, to identify new critical factors important for early osteoblastogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Fidgetin-like 1 gene inhibited by basic fibroblast growth factor regulates the proliferation and differentiation of osteoblasts.J Bone Miner Res. 2007 Jun;22(6):889-96. doi: 10.1359/jbmr.070311. J Bone Miner Res. 2007. PMID: 17352653
-
Regulation of osteoblast differentiation by Nurr1 in MC3T3-E1 cell line and mouse calvarial osteoblasts.J Cell Biochem. 2006 Oct 15;99(3):986-94. doi: 10.1002/jcb.20990. J Cell Biochem. 2006. PMID: 16741951
-
Identification of novel regulators associated with early-phase osteoblast differentiation.J Bone Miner Res. 2004 Jun;19(6):947-58. doi: 10.1359/JBMR.040216. Epub 2004 Feb 20. J Bone Miner Res. 2004. PMID: 15125793
-
Geranylgeranyl pyrophosphate stimulates PPARγ expression and adipogenesis through the inhibition of osteoblast differentiation.Bone. 2012 Feb;50(2):467-76. doi: 10.1016/j.bone.2011.09.056. Epub 2011 Oct 14. Bone. 2012. PMID: 22019459
-
Cellular and molecular effects of growth hormone and estrogen on human bone cells.APMIS Suppl. 1997;71:1-30. APMIS Suppl. 1997. PMID: 9357492 Review.
Cited by
-
Downregulation of the Autism Spectrum Disorder Gene Shank2 Decreases Bone Mass in Male Mice.JBMR Plus. 2022 Dec 15;7(2):e10711. doi: 10.1002/jbm4.10711. eCollection 2023 Feb. JBMR Plus. 2022. PMID: 36751416 Free PMC article.
-
Inhibition of Cdk5 Ameliorates Skeletal Bone Loss in Glucocorticoid-Treated Mice.Biomedicines. 2022 Feb 8;10(2):404. doi: 10.3390/biomedicines10020404. Biomedicines. 2022. PMID: 35203613 Free PMC article.
-
Impaired MC3T3-E1 osteoblast differentiation triggered by oncogenic HRAS is rescued by the farnesyltransferase inhibitor Tipifarnib.Sci Rep. 2025 Feb 26;15(1):6832. doi: 10.1038/s41598-025-91592-x. Sci Rep. 2025. PMID: 40000861 Free PMC article.
-
A variant in IL6ST with a selective IL-11 signaling defect in human and mouse.Bone Res. 2020 Jun 11;8:24. doi: 10.1038/s41413-020-0098-z. eCollection 2020. Bone Res. 2020. PMID: 32566365 Free PMC article.
-
Inhibition of Cdk5 increases osteoblast differentiation and bone mass and improves fracture healing.Bone Res. 2022 Apr 6;10(1):33. doi: 10.1038/s41413-022-00195-z. Bone Res. 2022. PMID: 35383146 Free PMC article.
References
-
- Qiang Y-W, et al. On the Molecular Mechanism of DKK1 Inhibition of Osteoblast Differentiation in Multiple Myeloma. Blood. 2006;108:3429–3429.
-
- Glorieux, F. H. Osteogenesis imperfecta. A disease of the osteoblast. Lancet (London, England)358 Suppl, S45 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

