The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units
- PMID: 30232462
- PMCID: PMC6145917
- DOI: 10.1038/s41598-018-32221-8
The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units
Abstract
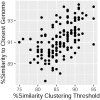
The complex community of microbes living in the human gut plays an important role in host wellbeing. However, defining a 'healthy' gut microbiome in terms of composition has remained an elusive task, despite its anticipated medical and scientific importance. In this regard, a central question has been if there is a 'core' microbiome consisting of bacterial groups common to all healthy humans. Recent studies have been able to define a compositional core in human gut microbiome datasets in terms of taxonomic assignments. However, the description of the core microbiome in terms of taxonomic assignments may not be adequate when considering subsequent analyses and applications. Through the implementation of a dynamic clustering approach in the meta-analyisis of comprehensive 16S rRNA marker gene datasets, this study found that the human gut pan-microbiome presents a preeminent compositional core comprised of discrete units of varying phylogenetic depth present in all individuals studied. Since both microbial traits and ecological coherence show signs of phylogenetic conservation, this outcome provides a new conceptual framework in the study of the ecosystem, as well as important practical considerations which should be taken into account in future research.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

