The Effect of Strain Level Diversity on Robust Inference of Virus-Induced Mortality of Phytoplankton
- PMID: 30233501
- PMCID: PMC6129610
- DOI: 10.3389/fmicb.2018.01850
The Effect of Strain Level Diversity on Robust Inference of Virus-Induced Mortality of Phytoplankton
Abstract
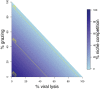
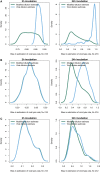
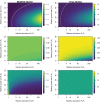
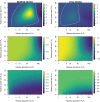
Infection and lysis of phytoplankton by viruses affects population dynamics and nutrient cycles within oceanic microbial communities. However, estimating the quantitative rates of viral-induced lysis remains challenging in situ. The modified dilution method is the most commonly utilized empirical approach to estimate virus-induced killing rates of phytoplankton. The lysis rate estimates of the modified dilution method are based on models which assume virus-host interactions can be represented by a single virus and a single host population with homogeneous life-history traits. Here, using modeling approaches, we examine the robustness of the modified dilution method in multi-strain, complex communities. We assume that strains differ in their life history traits, including growth rates (of hosts) and lysis rates (by viruses). We show that trait differences affect resulting experimental dynamics such that lysis rates measured using the modified dilution method may be driven by the fastest replicating strains; which are not necessarily the most abundant in situ. We discuss the implications of using the modified dilution method and alternative dilution-based approaches for estimating viral-induced lysis rates in marine microbial communities.
Keywords: dilution method; diversity; trait-based models; viral lysis; viruses.
Figures






References
-
- Arias A., Saiz E., Calbet A. (2017). Diel feeding rhythms in marine microzooplankton: effects of prey concentration, prey condition, and grazer nutritional history. Mar. Biol. 164:205 10.1007/s00227-017-3233-7 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

