Comparison of Highly and Weakly Virulent Dickeya solani Strains, With a View on the Pangenome and Panregulon of This Species
- PMID: 30233505
- PMCID: PMC6127512
- DOI: 10.3389/fmicb.2018.01940
Comparison of Highly and Weakly Virulent Dickeya solani Strains, With a View on the Pangenome and Panregulon of This Species
Abstract
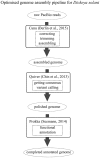
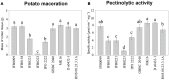
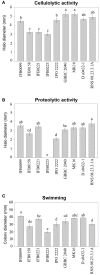
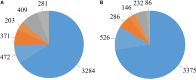
Bacteria belonging to the genera Dickeya and Pectobacterium are responsible for significant economic losses in a wide variety of crops and ornamentals. During last years, increasing losses in potato production have been attributed to the appearance of Dickeya solani. The D. solani strains investigated so far share genetic homogeneity, although different virulence levels were observed among strains of various origins. The purpose of this study was to investigate the genetic traits possibly related to the diverse virulence levels by means of comparative genomics. First, we developed a new genome assembly pipeline which allowed us to complete the D. solani genomes. Four de novo sequenced and ten publicly available genomes were used to identify the structure of the D. solani pangenome, in which 74.8 and 25.2% of genes were grouped into the core and dispensable genome, respectively. For D. solani panregulon analysis, we performed a binding site prediction for four transcription factors, namely CRP, KdgR, PecS and Fur, to detect the regulons of these virulence regulators. Most of the D. solani potential virulence factors were predicted to belong to the accessory regulons of CRP, KdgR, and PecS. Thus, some differences in gene expression could exist between D. solani strains. The comparison between a highly and a low virulent strain, IFB0099 and IFB0223, respectively, disclosed only small differences between their genomes but significant differences in the production of virulence factors like pectinases, cellulases and proteases, and in their mobility. The D. solani strains also diverge in the number and size of prophages present in their genomes. Another relevant difference is the disruption of the adhesin gene fhaB2 in the highly virulent strain. Strain IFB0223, which has a complete adhesin gene, is less mobile and less aggressive than IFB0099. This suggests that in this case, mobility rather than adherence is needed in order to trigger disease symptoms. This study highlights the utility of comparative genomics in predicting D. solani traits involved in the aggressiveness of this emerging plant pathogen.
Keywords: Pectobacteriaceae; adhesin; genome comparison; prophages; regulon comparison.
Figures



References
-
- Adeolu M., Alnajar S., Naushad S., Gupta R. (2016). Genome-based phylogeny and taxonomy of the “Enterobacteriales”: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
-
- Bailey T. L., Elkan C. (1994). Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. UCSD Technical Report CS94-351. Toronto, ON: University of California San Diego (UCSD). - PubMed
-
- Barras F., van Gijsegem F., Chatterjee A. K. (1994). Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32, 201–234. 10.1146/annurev.py.32.090194.001221 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

