Whole Genome Characterization of a Few EMS-Induced Mutants of Upland Rice Variety Nagina 22 Reveals a Staggeringly High Frequency of SNPs Which Show High Phenotypic Plasticity Towards the Wild-Type
- PMID: 30233603
- PMCID: PMC6132179
- DOI: 10.3389/fpls.2018.01179
Whole Genome Characterization of a Few EMS-Induced Mutants of Upland Rice Variety Nagina 22 Reveals a Staggeringly High Frequency of SNPs Which Show High Phenotypic Plasticity Towards the Wild-Type
Abstract
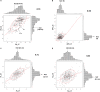
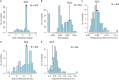
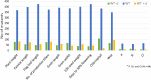
The Indian initiative, in creating mutant resources for the functional genomics in rice, has been instrumental in the development of 87,000 ethylmethanesulfonate (EMS)-induced mutants, of which 7,000 are in advanced generations. The mutants have been created in the background of Nagina 22, a popular drought- and heat-tolerant upland cultivar. As it is a pregreen revolution cultivar, as many as 573 dwarf mutants identified from this resource could be useful as an alternate source of dwarfing. A total of 541 mutants, including the macromutants and the trait-specific ones, obtained after appropriate screening, are being maintained in the mutant garden. Here, we report on the detailed characterizations of the 541 mutants based on the distinctness, uniformity, and stability (DUS) descriptors at two different locations. About 90% of the mutants were found to be similar to the wild type (WT) with high similarity index (>0.6) at both the locations. All 541 mutants were characterized for chlorophyll and epicuticular wax contents, while a subset of 84 mutants were characterized for their ionomes, namely, phosphorous, silicon, and chloride contents. Genotyping of these mutants with 54 genomewide simple sequence repeat (SSR) markers revealed 93% of the mutants to be either completely identical to WT or nearly identical with just one polymorphic locus. Whole genome resequencing (WGS) of four mutants, which have minimal differences in the SSR fingerprint pattern and DUS characters from the WT, revealed a staggeringly high number of single nucleotide polymorphisms (SNPs) on an average (16,453 per mutant) in the genic sequences. Of these, nearly 50% of the SNPs led to non-synonymous codons, while 30% resulted in synonymous codons. The number of insertions and deletions (InDels) varied from 898 to 2,595, with more than 80% of them being 1-2 bp long. Such a high number of SNPs could pose a serious challenge in identifying gene(s) governing the mutant phenotype by next generation sequencing-based mapping approaches such as Mutmap. From the WGS data of the WT and the mutants, we developed a genic resource of the WT with a novel analysis pipeline. The entire information about this resource along with the panicle architecture of the 493 mutants is made available in a mutant database EMSgardeN22 (http://14.139.229.201/EMSgardeN22).
Keywords: EMS mutants; Nagina 22; ionomics; mutant resource; mutation frequency; rice.
Figures





References
-
- Amitha Mithra S. V., Kar M. K., Mohapatra T., Robin S., Sarla N., Seshashayee M., et al. (2016). DBT propelled national effort in creating mutant resource for functional genomics in rice. Curr. Sci. 110 543–548. 10.18520/cs/v110/i4/543-548 - DOI
-
- Cantos C., Francisco P., Trijatmiko K. R., Slamet-Loedin I., Chadha-Mohanty P. K. (2014). Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front. Plant Sci. 5:302. 10.3389/fpls.2014.00302 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

