Quantification of Intrinsically Disordered Proteins: A Problem Not Fully Appreciated
- PMID: 30234128
- PMCID: PMC6131523
- DOI: 10.3389/fmolb.2018.00083
Quantification of Intrinsically Disordered Proteins: A Problem Not Fully Appreciated
Abstract
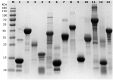
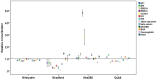
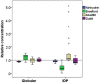
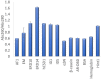
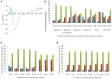
Protein quantification is essential in a great variety of biochemical assays, yet the inherent systematic errors associated with the concentration determination of intrinsically disordered proteins (IDPs) using classical methods are hardly appreciated. Routinely used assays for protein quantification, such as the Bradford assay or ultraviolet absorbance at 280 nm, usually seriously misestimate the concentrations of IDPs due to their distinct and variable amino acid composition. Therefore, dependable method(s) have to be worked out/adopted for this task. By comparison to elemental analysis as the gold standard, we show through the example of four globular proteins and nine IDPs that the ninhydrin assay and the commercial QubitTM Protein Assay provide reliable data on IDP quantity. However, as IDPs can show extreme variation in amino acid composition and physical features not necessarily covered by our examples, even these techniques should only be used for IDPs following standardization. The far-reaching implications of these simple observations are demonstrated through two examples: (i) circular dichroism spectrum deconvolution, and (ii) receptor-ligand affinity determination. These actual comparative examples illustrate the potential errors that can be incorporated into the biophysical parameters of IDPs, due to systematic misestimation of their concentration. This leads to inaccurate description of IDP functions.
Keywords: UV absorbance; circular dichroism; coomassie brilliant blue; elemental analysis; error propagation; nanoorange; ninhydrin; protein concentration.
Figures
References
-
- Calderon-Celis F., Diez-Fernandez S., Costa-Fernandez J. M., Encinar J. R., Calvete J. J., Sanz-Medel A. (2016). Elemental mass spectrometry for absolute intact protein quantification without protein-specific standards: application to snake venomics. Anal. Chem. 88, 9699–9706. 10.1021/acs.analchem.6b02585 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

