Pseudomonas aeruginosa displays a dormancy phenotype during long-term survival in water
- PMID: 30235203
- PMCID: PMC6147739
- DOI: 10.1371/journal.pone.0198384
Pseudomonas aeruginosa displays a dormancy phenotype during long-term survival in water
Abstract
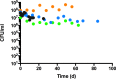
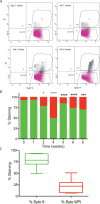
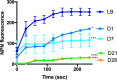
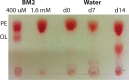
Pseudomonas aeruginosa is capable of long-term survival in water, which may serve as a reservoir for infection. Although viable cell counts of PAO1 incubated in water remain stable throughout 8 weeks, LIVE/DEAD staining indicated a high proportion of cells stained with propidium iodide (PI). The proportion of PI-stained cells increased by 4 weeks, then decreased again by 8 weeks, suggesting an adaptive response. This was also evident in an observed shift in cell morphology from a rod to a coccoid shape after 8 weeks. Fluorescence-activated cell sorting (FACS) was used to recover PI-stained cells, which were plated and shown to be viable, indicating that PI-stained cells were membrane-compromised but still cultivable. PAO1 mid-log cells in water were labeled with the dsDNA-binding dye PicoGreen to monitor viability as well as DNA integrity, which demonstrated that the population remains viable and transitions towards increased dsDNA staining. Metabolic activity was found to decrease significantly in water by 4 weeks. The PAO1 outer membrane became less permeable and more resistant to polymyxin B damage in water, and the profile of total membrane lipids changed over time. Among the ~1400 transcriptional lux fusions, gene expression in water revealed that the majority of genes were repressed, but subsets of genes were induced at particular time points. In summary, these results indicate that P. aeruginosa is dormant in water and this adaptation involves a complex pattern of gene regulation and changes to the cell to promote long-term survival and antibiotic tolerance. The approach of P. aeruginosa incubated in water may be useful to study antibiotic tolerance and the mechanisms of dormancy and survival in nutrient limiting conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67(3);351–368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

