The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae
- PMID: 30235899
- PMCID: PMC6162548
- DOI: 10.3390/genes9090462
The GIS2 Gene Is Repressed by a Zinc-Regulated Bicistronic RNA in Saccharomyces cerevisiae
Abstract
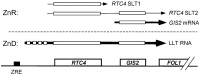
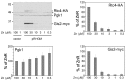
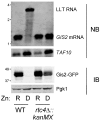
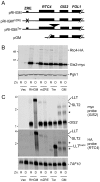
Zinc homeostasis is essential for all organisms. The Zap1 transcriptional activator regulates these processes in the yeast Saccharomyces cerevisiae. During zinc deficiency, Zap1 increases expression of zinc transporters and proteins involved in adapting to the stress of zinc deficiency. Transcriptional activation by Zap1 can also repress expression of some genes, e.g., RTC4. In zinc-replete cells, RTC4 mRNA is produced with a short transcript leader that is efficiently translated. During deficiency, Zap1-dependent expression of an RNA with a longer transcript leader represses the RTC4 promoter. This long leader transcript (LLT) is not translated due to the presence of small open reading frames upstream of the RTC4 coding region. In this study, we show that the RTC4 LLT RNA also plays a second function, i.e., repression of the adjacent GIS2 gene. In generating the LLT transcript, RNA polymerase II transcribes RTC4 through the GIS2 promoter. Production of the LLT RNA correlates with the decreased expression of GIS2 mRNA and mutations that prevent synthesis of the LLT RNA or terminate it before the GIS2 promoter renders GIS2 mRNA expression and Gis2 protein accumulation constitutive. Thus, we have discovered an unusual regulatory mechanism that uses a bicistronic RNA to control two genes simultaneously.
Keywords: RNA; bicistronic; gene expression; homeostasis; regulation; transcription; zinc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

