Predicting polymorphism in molecular crystals using orientational entropy
- PMID: 30237287
- PMCID: PMC6187181
- DOI: 10.1073/pnas.1811056115
Predicting polymorphism in molecular crystals using orientational entropy
Abstract
We introduce a computational method to discover polymorphs in molecular crystals at finite temperature. The method is based on reproducing the crystallization process starting from the liquid and letting the system discover the relevant polymorphs. This idea, however, conflicts with the fact that crystallization has a timescale much longer than that of molecular simulations. To bring the process within affordable simulation time, we enhance the fluctuations of a collective variable by constructing a bias potential with well-tempered metadynamics. We use as a collective variable an entropy surrogate based on an extended pair correlation function that includes the correlation between the orientations of pairs of molecules. We also propose a similarity metric between configurations based on the extended pair correlation function and a generalized Kullback-Leibler divergence. In this way, we automatically classify the configurations as belonging to a given polymorph, using our metric and a hierarchical clustering algorithm. We apply our method to urea and naphthalene. We find different polymorphs for both substances, and one of them is stabilized at finite temperature by entropic effects.
Keywords: crystal structure prediction; enhanced sampling; molecular simulation; polymorphism; urea.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
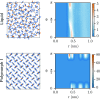

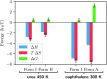
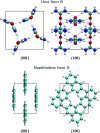
Similar articles
-
Enhancing Entropy and Enthalpy Fluctuations to Drive Crystallization in Atomistic Simulations.Phys Rev Lett. 2017 Jul 7;119(1):015701. doi: 10.1103/PhysRevLett.119.015701. Epub 2017 Jul 6. Phys Rev Lett. 2017. PMID: 28731736
-
Generating Cocrystal Polymorphs with Information Entropy Driven by Molecular Dynamics-Based Enhanced Sampling.J Phys Chem Lett. 2020 Nov 19;11(22):9751-9758. doi: 10.1021/acs.jpclett.0c02647. Epub 2020 Nov 3. J Phys Chem Lett. 2020. PMID: 33141590
-
Molecular-dynamics simulations of urea nucleation from aqueous solution.Proc Natl Acad Sci U S A. 2015 Jan 6;112(1):E6-14. doi: 10.1073/pnas.1421192111. Epub 2014 Dec 9. Proc Natl Acad Sci U S A. 2015. PMID: 25492932 Free PMC article.
-
Recent progress of structural study of polymorphic pharmaceutical drugs.Adv Drug Deliv Rev. 2017 Aug 1;117:71-85. doi: 10.1016/j.addr.2016.12.001. Epub 2016 Dec 7. Adv Drug Deliv Rev. 2017. PMID: 27940141 Review.
-
Collective Variables for Crystallization Simulations-from Early Developments to Recent Advances.ACS Omega. 2022 Dec 27;8(1):127-146. doi: 10.1021/acsomega.2c06310. eCollection 2023 Jan 10. ACS Omega. 2022. PMID: 36643553 Free PMC article. Review.
Cited by
-
Stochastic Resetting for Enhanced Sampling.J Phys Chem Lett. 2022 Dec 8;13(48):11230-11236. doi: 10.1021/acs.jpclett.2c03055. Epub 2022 Nov 29. J Phys Chem Lett. 2022. PMID: 36446130 Free PMC article.
-
Exploration vs Convergence Speed in Adaptive-Bias Enhanced Sampling.J Chem Theory Comput. 2022 Jun 14;18(6):3988-3996. doi: 10.1021/acs.jctc.2c00152. Epub 2022 May 26. J Chem Theory Comput. 2022. PMID: 35617155 Free PMC article.
-
Collective Variables for Conformational Polymorphism in Molecular Crystals.J Phys Chem Lett. 2023 Feb 2;14(4):971-976. doi: 10.1021/acs.jpclett.2c03491. Epub 2023 Jan 23. J Phys Chem Lett. 2023. PMID: 36689770 Free PMC article.
-
Enantiotropy of Simvastatin as a Result of Weakened Interactions in the Crystal Lattice: Entropy-Driven Double Transitions and the Transient Modulated Phase as Seen by Solid-State NMR Spectroscopy.Molecules. 2022 Jan 20;27(3):679. doi: 10.3390/molecules27030679. Molecules. 2022. PMID: 35163943 Free PMC article.
-
Machine-Learned Free Energy Surfaces for Capillary Condensation and Evaporation in Mesopores.Entropy (Basel). 2022 Jan 7;24(1):97. doi: 10.3390/e24010097. Entropy (Basel). 2022. PMID: 35052123 Free PMC article.
References
-
- Bernstein J. Polymorphism in Molecular Crystals. Vol 14 Oxford Univ Press; New York: 2002.
-
- Hilfiker R. Polymorphism: In the Pharmaceutical Industry. Wiley; Weinheim, Germany: 2006.
-
- Cabri W, Ghetti P, Pozzi G, Alpegiani M. Polymorphisms and patent, market, and legal battles: Cefdinir case study. Org Process Res Dev. 2007;11:64–72.
-
- Bauer J, et al. Ritonavir: An extraordinary example of conformational polymorphism. Pharm Res. 2001;18:859–866. - PubMed
-
- Bazterra VE, Ferraro MB, Facelli JC. Modified genetic algorithm to model crystal structures. I. Benzene, naphthalene and anthracene. J Chem Phys. 2002;116:5984–5991.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

