CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method
- PMID: 30237405
- PMCID: PMC6148268
- DOI: 10.1038/s41598-018-32329-x
CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method
Abstract
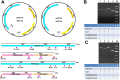
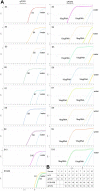
This study develops a new method for detecting and typing target DNA based on Cas9 nuclease, which was named as ctPCR, representing Cas9-sgRNA- or CRISPR-typing PCR. The technique can detect and type target DNA easily, rapidly, specifically, and sensitively. This technique detects target DNA in three steps: (1) amplifying target DNA with PCR by using a pair of universal primers (PCR1); (2) treating PCR1 products with a process referred to as CAT, representing Cas9 cutting, A tailing and T adaptor ligation; (3) amplifying the CAT-treated DNA with PCR by using a pair of general-specific primers (gs-primers) (PCR2). This method was verified by detecting HPV16 and HPV18 L1 gene in 13 different high-risk human papillomavirus (HPV) subtypes. This method was also verified by detecting the L1 and E6-E7 genes of two high-risk HPVs (HPV16 and 18) in cervical carcinoma cells and many clinical samples. In this method, PCR1 was performed to determine if the detected DNA sample contained the target DNA (such as virus infection), while PCR2 was performed to discriminate which genotypic target DNA was present in the detected DNA sample (such as virus subtypes). Based on these proof-of-concept experiments, this study provides a new CRISPR/Cas9-based DNA detection and typing method.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique.Anal Bioanal Chem. 2018 May;410(12):2889-2900. doi: 10.1007/s00216-018-0873-5. Epub 2018 Mar 15. Anal Bioanal Chem. 2018. PMID: 29546544
-
Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique.Anal Biochem. 2018 Nov 15;561-562:37-46. doi: 10.1016/j.ab.2018.09.012. Epub 2018 Sep 19. Anal Biochem. 2018. PMID: 30243976
-
A One-Pot CRISPR/Cas9-Typing PCR for DNA Detection and Genotyping.J Mol Diagn. 2021 Jan;23(1):46-60. doi: 10.1016/j.jmoldx.2020.10.004. Epub 2020 Oct 27. J Mol Diagn. 2021. PMID: 33127524
-
Oncogenic Human Papillomavirus: Application of CRISPR/Cas9 Therapeutic Strategies for Cervical Cancer.Cell Physiol Biochem. 2017;44(6):2455-2466. doi: 10.1159/000486168. Epub 2017 Dec 18. Cell Physiol Biochem. 2017. PMID: 29268281 Review.
-
The adaptive bacterial immune system CRISPR-Cas and its therapeutic potential.Med Monatsschr Pharm. 2017 Jan;40(1):17-23. Med Monatsschr Pharm. 2017. PMID: 29952526 Review. English, German.
Cited by
-
CRISPR-Cas systems for diagnosing infectious diseases.Methods. 2022 Jul;203:431-446. doi: 10.1016/j.ymeth.2021.04.007. Epub 2021 Apr 9. Methods. 2022. PMID: 33839288 Free PMC article. Review.
-
Description of CRISPR-Cas9 development and its prospects in human papillomavirus-driven cancer treatment.Front Immunol. 2022 Nov 21;13:1037124. doi: 10.3389/fimmu.2022.1037124. eCollection 2022. Front Immunol. 2022. PMID: 36479105 Free PMC article. Review.
-
PCR past, present and future.Biotechniques. 2020 Oct;69(4):317-325. doi: 10.2144/btn-2020-0057. Epub 2020 Aug 20. Biotechniques. 2020. PMID: 32815744 Free PMC article. Review.
-
CRISPR-Cas-Integrated LAMP.Biosensors (Basel). 2022 Nov 17;12(11):1035. doi: 10.3390/bios12111035. Biosensors (Basel). 2022. PMID: 36421156 Free PMC article. Review.
-
Human papilloma virus (HPV) mediated cancers: an insightful update.J Transl Med. 2025 Apr 29;23(1):483. doi: 10.1186/s12967-025-06470-x. J Transl Med. 2025. PMID: 40301924 Free PMC article. Review.
References
-
- Zhang K, Lin G, Li J. Quantitative nucleic acid amplification by digital PCR for clinical viral diagnostics. Clin. Chem. Lab. Med. 2016;54:1427–1433. - PubMed
-
- Behzadi P, Behzadi E, Alavian SM. DNA microarray technology in HBV genotyping. Minerva medica. 2017;108:473–476. - PubMed
-
- Han GY, et al. Detection of hereditary hearing loss gene by DNA microarray. Eur. rev. med. pharmaco. sci. 2017;21:3538–3542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

