Robust single-cell DNA methylome profiling with snmC-seq2
- PMID: 30237449
- PMCID: PMC6147798
- DOI: 10.1038/s41467-018-06355-2
Robust single-cell DNA methylome profiling with snmC-seq2
Abstract
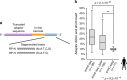
Single-cell DNA methylome profiling has enabled the study of epigenomic heterogeneity in complex tissues and during cellular reprogramming. However, broader applications of the method have been impeded by the modest quality of sequencing libraries. Here we report snmC-seq2, which provides improved read mapping, reduced artifactual reads, enhanced throughput, as well as increased library complexity and coverage uniformity compared to snmC-seq. snmC-seq2 is an efficient strategy suited for large-scale single-cell epigenomic studies.
Conflict of interest statement
L.K. is an inventor on a patent application (US 14/384,113) submitted by Swift Biosciences Inc. that covers Adaptase. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
- R21 MH112161/MH/NIMH NIH HHS/United States
- 1R21HG009274/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)/International
- 5U01MH105985/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)/International
- R21 HG009274/HG/NHGRI NIH HHS/United States
- U01 MH105985/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

