Identification of tRNA-derived small noncoding RNAs as potential biomarkers for prediction of recurrence in triple-negative breast cancer
- PMID: 30239174
- PMCID: PMC6198211
- DOI: 10.1002/cam4.1761
Identification of tRNA-derived small noncoding RNAs as potential biomarkers for prediction of recurrence in triple-negative breast cancer
Abstract
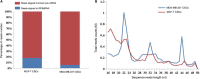
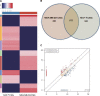
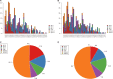
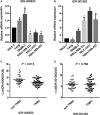
Triple-negative breast cancer (TNBC), an intrinsic subtype of breast cancer, is characterized by aggressive pathology and shorter overall survival. Yet there is no effective therapy for these patients. Breast cancer stem cells (BCSCs) in TNBC may account for treatment failure. It is urgent to identify new therapeutic targets for TNBC. tRNA-derived small noncoding RNAs (tDRs) represent a new class of small noncoding RNAs (sncRNA), which have been reported in some human diseases and biological processes. However, there is no detailed information about the relationship between tDRs and BCSCs. In this study, a population of CD44+ /CD24-/low cells was isolated and identified by reliable BCSC surface markers. tDR expression profiles in TNBC and non-TNBC CSCs were performed by RNA sequencing. A total of 1327 differentially expressed tDRs contained 18 upregulated and 54 downregulated in TNBC group. Furthermore, the selected tDRs were validated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). tDR-000620 expression level was consistently lower in TNBC cell lines CSCs (all P < 0.05) and serum samples (t = 2.597, P = 0.013). tDR-000620 expression was significant association with age (P = 0.018), node status (P = 0.026) and local recurrence (P = 0.019) by chi-square test. Kaplan-Meier method with log-rank test for comparison of recurrence curves. The results showed that the tDR-000620 expression (P = 0.002) and node status (P = 0.001) group were statistically significant with recurrence-free survival. Furthermore, multivariate Cox regression demonstrated that lymphatic metastasis (HR, 3.616; 95% CI, 1.234-10.596; P = 0.019) and low tDR-000620 expression (HR, 0.265; 95% CI, 0.073-0.959; P = 0.043) were two independent adverse predictive factors for recurrence-free survival. Finally, we found that tDR-000620 participated in some important biological processes though GO and KEGG analysis. Taken together, our study reveals the expression profiles of tDRs in TNBC and non-TNBC CSCs. It offers helpful information to understand the tDR-000620 expression is responsible for the aggressive phenotype of BCSCs. It may provide predictive biomarkers and therapeutic targets for TNBC recurrence.
Keywords: biomarker; stem cells; tDRs; triple-negative breast cancer.
© 2018 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures


References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7‐30. - PubMed
-
- Carey L, Winer E, Viale G, et al. Triple‐negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7(12):683‐692. - PubMed
-
- Agarwal G, Nanda G, Lal P, et al. Outcomes of triple‐negative breast cancers (TNBC) compared with non‐TNBC: does the survival vary for all stages? World J Surg. 2016;40(6):1362‐1372. - PubMed
-
- Mackillop WJ, Ciampi A, Till JE, et al. A stem cell model of human tumor growth: implications for tumor cell clonogenic assays. J Natl Cancer Inst. 1983;70(1):9‐16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

