Pulmonary Metagenomic Sequencing Suggests Missed Infections in Immunocompromised Children
- PMID: 30239621
- PMCID: PMC6784263
- DOI: 10.1093/cid/ciy802
Pulmonary Metagenomic Sequencing Suggests Missed Infections in Immunocompromised Children
Abstract
Background: Despite improved diagnostics, pulmonary pathogens in immunocompromised children frequently evade detection, leading to significant mortality. Therefore, we aimed to develop a highly sensitive metagenomic next-generation sequencing (mNGS) assay capable of evaluating the pulmonary microbiome and identifying diverse pathogens in the lungs of immunocompromised children.
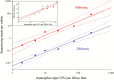
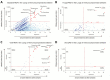
Methods: We collected 41 lower respiratory specimens from 34 immunocompromised children undergoing evaluation for pulmonary disease at 3 children's hospitals from 2014-2016. Samples underwent mechanical homogenization, parallel RNA/DNA extraction, and metagenomic sequencing. Sequencing reads were aligned to the National Center for Biotechnology Information nucleotide reference database to determine taxonomic identities. Statistical outliers were determined based on abundance within each sample and relative to other samples in the cohort.
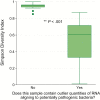
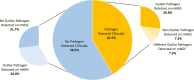
Results: We identified a rich cross-domain pulmonary microbiome that contained bacteria, fungi, RNA viruses, and DNA viruses in each patient. Potentially pathogenic bacteria were ubiquitous among samples but could be distinguished as possible causes of disease by parsing for outlier organisms. Samples with bacterial outliers had significantly depressed alpha-diversity (median, 0.61; interquartile range [IQR], 0.33-0.72 vs median, 0.96; IQR, 0.94-0.96; P < .001). Potential pathogens were detected in half of samples previously negative by clinical diagnostics, demonstrating increased sensitivity for missed pulmonary pathogens (P < .001).
Conclusions: An optimized mNGS assay for pulmonary microbes demonstrates significant inoculation of the lower airways of immunocompromised children with diverse bacteria, fungi, and viruses. Potential pathogens can be identified based on absolute and relative abundance. Ongoing investigation is needed to determine the pathogenic significance of outlier microbes in the lungs of immunocompromised children with pulmonary disease.
Keywords: immunocompromised host; intensive care units; metagenomics; microbiota; pediatric; respiratory tract infections.
© The Author(s) 2018. Published by Oxford University Press for the Infectious Diseases Society of America. All rights reserved. For permissions, e-mail: journals.permissions@oup.com.
Figures
References
-
- Center for International Blood and Marrow Transplant, a contractor for the C.W. Bill Young Cell Transplantation Program operated through the U. S. Department of Health and Human Services, Health Resources and Services Administration, Healthcare Systems Bureau. U.S. transplant data by age, number of transplants reported in 2016. Available at: https://bloodcell.transplant.hrsa.gov/research/transplant_data/transplan.... Accessed 1 July 2017.
-
- OPTN/SRTR 2015 annual data report: introduction. Am J Transplant 2017; 17(Suppl 1):11–20. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin 2017; 67:7–30. - PubMed
-
- Kaya Z, Weiner DJ, Yilmaz D, Rowan J, Goyal RK. Lung function, pulmonary complications, and mortality after allogeneic blood and marrow transplantation in children. Biol Blood Marrow Transplant 2009; 15:817–26. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

