Highly Selective Transmission Success of Dengue Virus Type 1 Lineages in a Dynamic Virus Population: An Evolutionary and Fitness Perspective
- PMID: 30240624
- PMCID: PMC6137288
- DOI: 10.1016/j.isci.2018.07.008
Highly Selective Transmission Success of Dengue Virus Type 1 Lineages in a Dynamic Virus Population: An Evolutionary and Fitness Perspective
Abstract
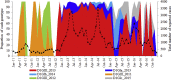
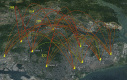
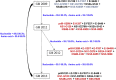
Arbovirus transmission is modulated by host, vector, virus, and environmental factors. Even though viral fitness plays a salient role in host and vector adaptation, the transmission success of individual strains in a heterogeneous population may be stochastic. Our large-scale molecular epidemiological analyses of a dengue virus type 1 population revealed that only a subset of strains (16.7%; n = 6) were able to sustain transmission, despite the population being widely dispersed, dynamic, and heterogeneous. The overall dominance was variable even among the "established" lineages, albeit sharing comparable evolutionary characteristics and replication profiles. These findings indicated that virological parameters alone were unlikely to have a profound effect on the survival of viral lineages, suggesting an important role for non-viral factors in the transmission success of lineages. Our observations, therefore, emphasize the strategic importance of a holistic understanding of vector, human host, and viral factors in the control of vector-borne diseases.
Keywords: Disease; Evolutionary Biology; Phylogenetics; Virology.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Genetic Drift, Purifying Selection and Vector Genotype Shape Dengue Virus Intra-host Genetic Diversity in Mosquitoes.PLoS Genet. 2016 Jun 15;12(6):e1006111. doi: 10.1371/journal.pgen.1006111. eCollection 2016 Jun. PLoS Genet. 2016. PMID: 27304978 Free PMC article.
-
Intra-epidemic evolutionary dynamics of a Dengue virus type 1 population reveal mutant spectra that correlate with disease transmission.Sci Rep. 2016 Mar 4;6:22592. doi: 10.1038/srep22592. Sci Rep. 2016. PMID: 26940650 Free PMC article.
-
Evolution of Subgenomic RNA Shapes Dengue Virus Adaptation and Epidemiological Fitness.iScience. 2019 Jun 28;16:94-105. doi: 10.1016/j.isci.2019.05.019. Epub 2019 May 16. iScience. 2019. PMID: 31154208 Free PMC article.
-
Molecular biological assessment methods and understanding the course of the HIV infection.APMIS Suppl. 2003;(114):1-37. APMIS Suppl. 2003. PMID: 14626050 Review.
-
The tortoise or the hare? Impacts of within-host dynamics on transmission success of arthropod-borne viruses.Philos Trans R Soc Lond B Biol Sci. 2015 Aug 19;370(1675):20140299. doi: 10.1098/rstb.2014.0299. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26150665 Free PMC article. Review.
Cited by
-
Molecular surveillance of arboviruses circulation and co-infection during a large chikungunya virus outbreak in Thailand, October 2018 to February 2020.Sci Rep. 2022 Dec 24;12(1):22323. doi: 10.1038/s41598-022-27028-7. Sci Rep. 2022. PMID: 36566236 Free PMC article.
-
Detection of Japanese Encephalitis Virus in Culex Mosquitoes in Singapore.Am J Trop Med Hyg. 2020 Sep;103(3):1234-1240. doi: 10.4269/ajtmh.19-0377. Am J Trop Med Hyg. 2020. PMID: 32700679 Free PMC article.
-
Lineage Replacement Associated with Fitness Gain in Mammalian Cells and Aedes aegypti: A Catalyst for Dengue Virus Type 2 Transmission.Microorganisms. 2022 May 26;10(6):1100. doi: 10.3390/microorganisms10061100. Microorganisms. 2022. PMID: 35744618 Free PMC article.
-
Novel and emerging mutations of SARS-CoV-2: Biomedical implications.Biomed Pharmacother. 2021 Jul;139:111599. doi: 10.1016/j.biopha.2021.111599. Epub 2021 Apr 23. Biomed Pharmacother. 2021. PMID: 33915502 Free PMC article. Review.
-
Genetic Diversity of Dengue Virus in Clinical Specimens from Bangkok, Thailand, during 2018-2020: Co-Circulation of All Four Serotypes with Multiple Genotypes and/or Clades.Trop Med Infect Dis. 2021 Sep 4;6(3):162. doi: 10.3390/tropicalmed6030162. Trop Med Infect Dis. 2021. PMID: 34564546 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

