Histone H2A.Z Suppression of Interferon-Stimulated Transcription and Antiviral Immunity Is Modulated by GCN5 and BRD2
- PMID: 30240626
- PMCID: PMC6137307
- DOI: 10.1016/j.isci.2018.07.013
Histone H2A.Z Suppression of Interferon-Stimulated Transcription and Antiviral Immunity Is Modulated by GCN5 and BRD2
Abstract
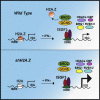
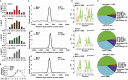
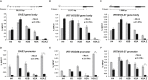
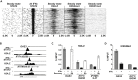
Type I interferon (IFN)-stimulated gene (ISG) expression requires interaction between a transcription factor complex, ISGF3, and target gene promoters to initiate transcription and protection against infection. To uncover chromatin regulatory features of this antiviral immune response, IFN-induced nucleosome and histone dynamics of human ISG loci were examined. ISGF3 recruitment after IFN stimulation was accompanied by nucleosome reorganization at promoters and gene bodies. IFN stimulation induced loss of core histones H2B, H3, and H4, as well as H2A.Z at ISG promoters. A strong correlation was found between H2A.Z occupancy and ISGF3 target sites, and IFN-stimulated H2A.Z removal requires STAT1, STAT2, and IRF9. Neither INO80 nor SWI/SNF participate in IFN-driven H2A.Z eviction, but GCN5 and BRD2 are required. Interference with H2A.Z expression enhanced ISGF3 recruitment to ISG promoters, ISG mRNA expression, and IFN-stimulated antiviral immunity. This indicates that H2A.Z nucleosomes at ISG promoters restrict optimal ISGF3 engagement and modulate the biological response to IFN.
Keywords: Immune Response; Immunology; Transcriptomics.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Aaronson D.S., Horvath C.M. A road map for those who don't know JAK-STAT. Science. 2002;296:1653–1655. - PubMed
-
- Banninger G., Reich N.C. STAT2 nuclear trafficking. J. Biol. Chem. 2004;279:39199–39206. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

