Marked Global DNA Hypomethylation Is Associated with Constitutive PD-L1 Expression in Melanoma
- PMID: 30240750
- PMCID: PMC6147024
- DOI: 10.1016/j.isci.2018.05.021
Marked Global DNA Hypomethylation Is Associated with Constitutive PD-L1 Expression in Melanoma
Abstract
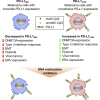
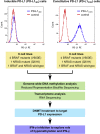
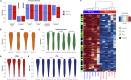
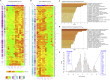
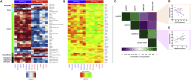
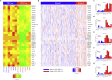
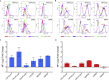
Constitutive expression of the immune checkpoint, PD-L1, inhibits anti-tumor immune responses in cancer, although the factors involved in PD-L1 regulation are poorly understood. Here we show that loss of global DNA methylation, particularly in intergenic regions and repeat elements, is associated with constitutive (PD-L1CON), versus inducible (PD-L1IND), PD-L1 expression in melanoma cell lines. We further show this is accompanied by transcriptomic up-regulation. De novo epigenetic regulators (e.g., DNMT3A) are strongly correlated with PD-L1 expression and methylome status. Accordingly, decitabine-mediated inhibition of global methylation in melanoma cells leads to increased PD-L1 expression. Moreover, viral mimicry and immune response genes are highly expressed in lymphocyte-negative plus PD-L1-positive melanomas, versus PD-L1-negative melanomas in The Cancer Genome Atlas (TCGA). In summary, using integrated genomic analysis we identified that global DNA methylation influences PD-L1 expression in melanoma, and hence melanoma's ability to evade anti-tumor immune responses. These results have implications for combining epigenetic therapy with immunotherapy.
Keywords: Cancer; Genetics; Genomics; Transcriptomics.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Aran D., Toperoff G., Rosenberg M., Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 2011;20:670–680. - PubMed
-
- Bird A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. - PubMed
-
- Chatterjee A., Macaulay E.C., Ahn A., Ludgate J.L., Stockwell P.A., Weeks R.J., Parry M.F., Foster T.J., Knarston I.M., Eccles M.R. Comparative assessment of DNA methylation patterns between reduced representation bisulfite sequencing and sequenom EpiTyper methylation analysis. Epigenomics. 2017;9:823–832. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

