ECPred: a tool for the prediction of the enzymatic functions of protein sequences based on the EC nomenclature
- PMID: 30241466
- PMCID: PMC6150975
- DOI: 10.1186/s12859-018-2368-y
ECPred: a tool for the prediction of the enzymatic functions of protein sequences based on the EC nomenclature
Abstract
Background: The automated prediction of the enzymatic functions of uncharacterized proteins is a crucial topic in bioinformatics. Although several methods and tools have been proposed to classify enzymes, most of these studies are limited to specific functional classes and levels of the Enzyme Commission (EC) number hierarchy. Besides, most of the previous methods incorporated only a single input feature type, which limits the applicability to the wide functional space. Here, we proposed a novel enzymatic function prediction tool, ECPred, based on ensemble of machine learning classifiers.
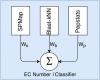
Results: In ECPred, each EC number constituted an individual class and therefore, had an independent learning model. Enzyme vs. non-enzyme classification is incorporated into ECPred along with a hierarchical prediction approach exploiting the tree structure of the EC nomenclature. ECPred provides predictions for 858 EC numbers in total including 6 main classes, 55 subclass classes, 163 sub-subclass classes and 634 substrate classes. The proposed method is tested and compared with the state-of-the-art enzyme function prediction tools by using independent temporal hold-out and no-Pfam datasets constructed during this study.
Conclusions: ECPred is presented both as a stand-alone and a web based tool to provide probabilistic enzymatic function predictions (at all five levels of EC) for uncharacterized protein sequences. Also, the datasets of this study will be a valuable resource for future benchmarking studies. ECPred is available for download, together with all of the datasets used in this study, at: https://github.com/cansyl/ECPred . ECPred webserver can be accessed through http://cansyl.metu.edu.tr/ECPred.html .
Keywords: Benchmark datasets; EC numbers; Function prediction; Machine learning; Protein sequence.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Cornish-Bowden A. Current IUBMB recommendations on enzyme nomenclature and kinetics. Perspect Sci. 2014;1:74–87. doi: 10.1016/j.pisc.2014.02.006. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

