Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations
- PMID: 30241495
- PMCID: PMC6151024
- DOI: 10.1186/s13059-018-1522-1
Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations
Abstract
Background: The genetic origins of Uralic speakers from across a vast territory in the temperate zone of North Eurasia have remained elusive. Previous studies have shown contrasting proportions of Eastern and Western Eurasian ancestry in their mitochondrial and Y chromosomal gene pools. While the maternal lineages reflect by and large the geographic background of a given Uralic-speaking population, the frequency of Y chromosomes of Eastern Eurasian origin is distinctively high among European Uralic speakers. The autosomal variation of Uralic speakers, however, has not yet been studied comprehensively.
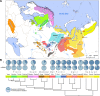
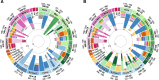
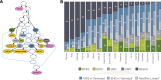
Results: Here, we present a genome-wide analysis of 15 Uralic-speaking populations which cover all main groups of the linguistic family. We show that contemporary Uralic speakers are genetically very similar to their local geographical neighbours. However, when studying relationships among geographically distant populations, we find that most of the Uralic speakers and some of their neighbours share a genetic component of possibly Siberian origin. Additionally, we show that most Uralic speakers share significantly more genomic segments identity-by-descent with each other than with geographically equidistant speakers of other languages. We find that correlated genome-wide genetic and lexical distances among Uralic speakers suggest co-dispersion of genes and languages. Yet, we do not find long-range genetic ties between Estonians and Hungarians with their linguistic sisters that would distinguish them from their non-Uralic-speaking neighbours.
Conclusions: We show that most Uralic speakers share a distinct ancestry component of likely Siberian origin, which suggests that the spread of Uralic languages involved at least some demic component.
Keywords: Genome-wide analysis; Haplotype analysis; IBD-segments; Population genetics; Uralic languages.
Conflict of interest statement
Ethics approval and consent to participate
DNA samples were obtained from unrelated volunteers, all donors provided informed consent and all experiments were performed in accordance with the relevant guidelines and regulations of the involved countries. The research has been approved by the Research Ethics Commitees of the University of Tartu and the Russian Academy of Sciences (approval nos. 228/M-40, 252/M-17, 17146-9217)
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Indreko R. Origin and area of settlement of the Fenno-Ugrian peoples. Science in Exile. Publication of the Scientific Quarterly “Scholar”. Heidelberg: Heidelberger Gutenberg-Druckerei GmbH; 1948. pp. 3–24.
-
- Setälä N. E (1926) Johdanto. In: Kannisto A, editor. Suomen suku I. Helsinki: Kustannusosakeyhtiö Otava.
Publication types
MeSH terms
Grants and funding
- PUT1217/Eesti Teadusagentuur/International
- PUT1339/Eesti Teadusagentuur/International
- IUT24/Eesti Teadusagentuur/International
- 2014-2020.4.01.15-0012/European Regional Development Fund/International
- 014-2020.4.01.16-0271/European Regional Development Fund/International
- 2014-2020.4.01.16-0125/European Regional Development Fund/International
- 0324-2018-0016/Russian Federation State Research Project/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- FP7-PEOPLE-2012-IRSES-number 318979/FP7 People: Marie-Curie Actions/International
- 16-06-00303/Russian Foundation for Basic Research/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

