Analysis of peptide-SLA binding by establishing immortalized porcine alveolar macrophage cells with different SLA class II haplotypes
- PMID: 30241566
- PMCID: PMC6151021
- DOI: 10.1186/s13567-018-0590-2
Analysis of peptide-SLA binding by establishing immortalized porcine alveolar macrophage cells with different SLA class II haplotypes
Abstract
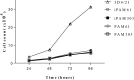
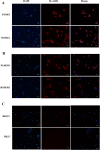
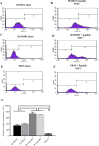
Primary porcine alveolar macrophages (PAM) are useful for studying viral infections and immune response in pigs; however, long-term use of these cells is limited by the cells' short lifespan. We immortalized primary PAMs by transfecting them with both hTERT and SV40LT and established two immortalized cell lines (iPAMs) actively proliferating even after 35 passages. These cells possessed the characteristics of primary PAMs, including strong expression of swine leukocyte antigen (SLA) class II genes and the inability to grow anchorage-independently. We characterized their SLA genes and subsequently performed peptide-SLA binding assays using a peptide from porcine circovirus type 2 open reading frame 2 to experimentally measure the binding affinity of the peptide to SLA class II. The number of peptides bound to cells measured by fluorescence was very low for PK15 cells (7.0% ± 1.5), which are not antigen-presenting cells, unlike iPAM61 (33.7% ± 3.4; SLA-DQA*0201/0303, DQB1*0201/0901, DRB1*0201/1301) and iPAM303 (73.3% ± 5.4; SLA DQA*0106/0201, DQB1*0202/0701, DRB1*0402/0602). The difference in peptide binding between the two iPAMs was likely due to the allelic differences between the SLA class II molecules that were expressed. The development of an immortal PAM cell panel harboring diverse SLA haplotypes and the use of an established method in this study can become a valuable tool for evaluating the interaction between antigenic peptides and SLA molecules and is important for many applications in veterinary medicine including vaccine development.
Figures


Similar articles
-
Development of an Immortalized Porcine Fibroblast Cell Panel With Different Swine Leukocyte Antigen Genotypes.Front Genet. 2022 Feb 7;13:815328. doi: 10.3389/fgene.2022.815328. eCollection 2022. Front Genet. 2022. PMID: 35198008 Free PMC article.
-
Molecular characterization of swine leukocyte antigen gene diversity in purebred Pietrain pigs.Anim Genet. 2013 Apr;44(2):202-5. doi: 10.1111/j.1365-2052.2012.02375.x. Epub 2012 May 16. Anim Genet. 2013. PMID: 22587706
-
Comparative analysis of swine leukocyte antigen gene diversity in European farmed pigs.Anim Genet. 2021 Aug;52(4):523-531. doi: 10.1111/age.13090. Epub 2021 May 24. Anim Genet. 2021. PMID: 34028065 Free PMC article.
-
Nomenclature for factors of the swine leukocyte antigen class II system, 2005.Tissue Antigens. 2005 Dec;66(6):623-39. doi: 10.1111/j.1399-0039.2005.00492.x. Tissue Antigens. 2005. PMID: 16305679 Review.
-
Molecular genetics of the swine major histocompatibility complex, the SLA complex.Dev Comp Immunol. 2009 Mar;33(3):362-74. doi: 10.1016/j.dci.2008.07.002. Epub 2008 Aug 27. Dev Comp Immunol. 2009. PMID: 18760302 Review.
Cited by
-
Establishment of an Immortalized Porcine Alveolar Macrophage Cell Line That Supports Efficient Replication of Porcine Reproductive and Respiratory Syndrome Viruses.Pathogens. 2024 Nov 21;13(12):1026. doi: 10.3390/pathogens13121026. Pathogens. 2024. PMID: 39770286 Free PMC article.
-
New Immunoinformatics Tools for Swine: Designing Epitope-Driven Vaccines, Predicting Vaccine Efficacy, and Making Vaccines on Demand.Front Immunol. 2020 Oct 5;11:563362. doi: 10.3389/fimmu.2020.563362. eCollection 2020. Front Immunol. 2020. PMID: 33123135 Free PMC article. Review.
-
A comparative analysis of SLA-DRB1 genetic diversity in Colombian (creoles and commercial line) and worldwide swine populations.Sci Rep. 2021 Feb 22;11(1):4340. doi: 10.1038/s41598-021-83637-8. Sci Rep. 2021. PMID: 33619347 Free PMC article.
-
Development of an Immortalized Porcine Fibroblast Cell Panel With Different Swine Leukocyte Antigen Genotypes.Front Genet. 2022 Feb 7;13:815328. doi: 10.3389/fgene.2022.815328. eCollection 2022. Front Genet. 2022. PMID: 35198008 Free PMC article.
-
Comparative Analysis of SLA-1, SLA-2, and DQB1 Genetic Diversity in Locally-Adapted Kenyan Pigs and Their Wild Relatives, Warthogs.Vet Sci. 2021 Sep 2;8(9):180. doi: 10.3390/vetsci8090180. Vet Sci. 2021. PMID: 34564574 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

