DNA Methylation Clocks in Aging: Categories, Causes, and Consequences
- PMID: 30241605
- PMCID: PMC6520108
- DOI: 10.1016/j.molcel.2018.08.008
DNA Methylation Clocks in Aging: Categories, Causes, and Consequences
Abstract
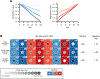
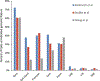
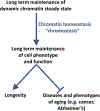
Age-associated changes to the mammalian DNA methylome are well documented and thought to promote diseases of aging, such as cancer. Recent studies have identified collections of individual methylation sites whose aggregate methylation status measures chronological age, referred to as the DNA methylation clock. DNA methylation may also have value as a biomarker of healthy versus unhealthy aging and disease risk; in other words, a biological clock. Here we consider the relationship between the chronological and biological clocks, their underlying mechanisms, potential consequences, and their utility as biomarkers and as targets for intervention to promote healthy aging and longevity.
Keywords: DNA methylation; aging; biological age; chronological age; clock; epigenetics.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures


References
-
- Ahuja N, Li Q, Mohan AL, Baylin SB, and Issa JP (1998). Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 58, 5489–5494. - PubMed
-
- Ambatipudi S, Horvath S, Perrier F, Cuenin C, Hernandez-Vargas H, Le Calvez-Kelm F, Durand G, Byrnes G, Ferrari P, Bouaoun L, et al. (2017). DNA methylome analysis identifies accelerated epigenetic ageing associated with postmenopausal breast cancer susceptibility. Eur. J. Cancer 75,299–307. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

